Systemic Homeostasis in Metabolome, Ionome, and Microbiome of Wild
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
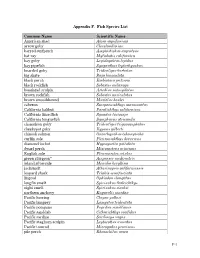
Appendix E: Fish Species List
Appendix F. Fish Species List Common Name Scientific Name American shad Alosa sapidissima arrow goby Clevelandia ios barred surfperch Amphistichus argenteus bat ray Myliobatis californica bay goby Lepidogobius lepidus bay pipefish Syngnathus leptorhynchus bearded goby Tridentiger barbatus big skate Raja binoculata black perch Embiotoca jacksoni black rockfish Sebastes melanops bonehead sculpin Artedius notospilotus brown rockfish Sebastes auriculatus brown smoothhound Mustelus henlei cabezon Scorpaenichthys marmoratus California halibut Paralichthys californicus California lizardfish Synodus lucioceps California tonguefish Symphurus atricauda chameleon goby Tridentiger trigonocephalus cheekspot goby Ilypnus gilberti chinook salmon Oncorhynchus tshawytscha curlfin sole Pleuronichthys decurrens diamond turbot Hypsopsetta guttulata dwarf perch Micrometrus minimus English sole Pleuronectes vetulus green sturgeon* Acipenser medirostris inland silverside Menidia beryllina jacksmelt Atherinopsis californiensis leopard shark Triakis semifasciata lingcod Ophiodon elongatus longfin smelt Spirinchus thaleichthys night smelt Spirinchus starksi northern anchovy Engraulis mordax Pacific herring Clupea pallasi Pacific lamprey Lampetra tridentata Pacific pompano Peprilus simillimus Pacific sanddab Citharichthys sordidus Pacific sardine Sardinops sagax Pacific staghorn sculpin Leptocottus armatus Pacific tomcod Microgadus proximus pile perch Rhacochilus vacca F-1 plainfin midshipman Porichthys notatus rainwater killifish Lucania parva river lamprey Lampetra -

Report on the Monitoring of Radionuclides in Fishery Products (March 2011 - January 2015)
Report on the Monitoring of Radionuclides in Fishery Products (March 2011 - January 2015) April 2015 Fisheries Agency of Japan 0 1 Table of Contents Overview…………………………………………………………………………………………………. 8 The Purpose of this Report………………………………………………………………………………9 Part One. Efforts to Guarantee the Safety of Fishery Products………………………………………..11 Chapter 1. Monitoring of Radioactive Materials in Food; Restrictions on Distribution and Other Countermeasures………...…………………………………………………………………11 1-1-1 Standard Limits for Radioactive Materials in Food………………………………………...……11 1-1-2 Methods of Testing for Radioactive Materials………………………………………...…………12 1-1-3 Inspections of Fishery Products for Radioactive Materials…………………………...…………14 1-1-4 Restrictions and Suspensions on Distribution and Shipping ……………………………………..18 1-1-5 Cancellation of Restrictions on Shipping and Distribution………………………………………20 Box 1 Calculation of the Limits for Human Consumption……..………………………………………23 Box 2 Survey of Radiation Dose from Radionuclides in Foods Calculation of the Limits…………….24 Box 3 Examples of Local Government Monitoring Plan………………………………...…………….25 Chapter 2. Results of Radioactive Cesium Inspections for Fishery Products…………………………26 1-2-1 Inspection Results for Nationwide Fishery Products in Japan (in total)…………………………26 1-2-2 Inspection Results for Fukushima Prefecture Fishery Products (all)…………………………….27 1-2-3 Inspection Results for Fishery Products (all) from Outside Fukushima Prefecture……………...30 1-2-4 Trends within Fish Species……………………………………………………………………….32 1-2-5 Inspection Results for Main Target Fish Species of Fishing and Farming by Fiscal Year……….42 1-2-6 Radioactive Material Concentrations within Fish within 20 km of the Fukushima Daiichi NPS.46 Box 4 Fukushima Fishing Trials………………………………...……………………………………...47 1-2-7 Screening Test by Prefectural and Municipal Governments……………………………………..48 Chapter 3. Inspection for Radionuclides Other Than Radioactive Cesium……………………………49 1-3-1 Inspections for Radioactive Strontium etc. -

A Dissertation Entitled Evolution, Systematics
A Dissertation Entitled Evolution, systematics, and phylogeography of Ponto-Caspian gobies (Benthophilinae: Gobiidae: Teleostei) By Matthew E. Neilson Submitted as partial fulfillment of the requirements for The Doctor of Philosophy Degree in Biology (Ecology) ____________________________________ Adviser: Dr. Carol A. Stepien ____________________________________ Committee Member: Dr. Christine M. Mayer ____________________________________ Committee Member: Dr. Elliot J. Tramer ____________________________________ Committee Member: Dr. David J. Jude ____________________________________ Committee Member: Dr. Juan L. Bouzat ____________________________________ College of Graduate Studies The University of Toledo December 2009 Copyright © 2009 This document is copyrighted material. Under copyright law, no parts of this document may be reproduced without the expressed permission of the author. _______________________________________________________________________ An Abstract of Evolution, systematics, and phylogeography of Ponto-Caspian gobies (Benthophilinae: Gobiidae: Teleostei) Matthew E. Neilson Submitted as partial fulfillment of the requirements for The Doctor of Philosophy Degree in Biology (Ecology) The University of Toledo December 2009 The study of biodiversity, at multiple hierarchical levels, provides insight into the evolutionary history of taxa and provides a framework for understanding patterns in ecology. This is especially poignant in invasion biology, where the prevalence of invasiveness in certain taxonomic groups could -

Inland Fishes of California
Inland Fishes of California Revise d and Expanded PETER B. MO YL E Illustrations by Chris Ma ri van Dyck and Joe Tome ller i NIVERS ITyor ALfFORNJA PRESS Ikrkd cr I.", ..\ n~d e ' Lon don Universit }, 0 Ca lifornia Press Herkdey and Los Angeles, Ca lifornia Uni ve rsity of alifornia Press, Ltd. Lundun, England ~ 2002 by the Regents of the Unive rsi ty of Ca lifornia Library of Cungress ataloging-in -Publ ica tion Data j\·[oyk, Pen: r B. Inland fis hes of California / Peter B. Moyle ; illustrations by Chris Mari van D)'ck and Joe Tomell eri.- Rev. and expanded. p. cm. In lu de> bibl iographical refe rences (p. ). ISBN 0- 20-2.2754 -'1 (cl ot h: alk. papa) I. rreshw:ltcr lishes-Cali(ornia. I. Title. QL62S C2 M6H 2002 597 .17/i'097Q4-dc21 20010 27680 1\!UlIl.Ifaclu rcd in Canacla II 10 Q9 00 07 06 0 04 03 02 10 ' 1\ 7 b '; -\ 3 2 1 Th paper u!)ed in thi> public.ltiu(] 111l'd., the minimum requirements "fA SI / i': ISO Z39.4H-1992 (R 199;) ( Pmlllllh'/l e ofPa pcr) . e Special Thanks The illustrations for this book were made possible by gra nts from the following : California-Nevada Chapter, American Fi she ries Soc iety Western Di vision, Am erican Fi she ries Society California Department of Fish and Game Giles W. and El ise G. Mead Foundation We appreciate the generous funding support toward the publication of this book by the United Sta tes Environme ntal Protection Agency, Region IX, San Francisco Contents Pre(acc ix Salmon and Trout, Salmonidae 242 Ackl10 11'ledgl11 el1ts Xlll Silversides, Atherinopsidae 307 COlll'er,<iol1 ['actors xv -

Japan Update to 05.04.2021 Approval No Name Address Products Number FROZEN CHUM SALMON DRESSED (Oncorhynchus Keta)
Japan Update to 05.04.2021 Approval No Name Address Products Number FROZEN CHUM SALMON DRESSED (Oncorhynchus keta). FROZEN DOLPHINFISH DRESSED (Coryphaena hippurus). FROZEN JAPANESE SARDINE ROUND (Sardinops 81,Misaki-Cho,Rausu- Kaneshin Tsuyama melanostictus). FROZEN ALASKA POLLACK DRESSED (Theragra chalcogramma). 1 VN01870001 Cho, Menashi- Co.,Ltd FROZEN ALASKA POLLACK ROUND (Theragra chalcogramma). FROZEN PACIFIC COD Gun,Hokkaido,Japan DRESSED. (Gadus macrocephalus). FROZEN PACIFIC COD ROUND. (Gadus macrocephalus) Maekawa Shouten Hokkaido Nemuro City Fresh Fish (Excluding Fish By-Product); Fresh Bivalve Mollusk.; Frozen Fish (Excluding 2 VN01860002 Co., Ltd Nishihamacho 10-177 Fish By-Product); Frozen Processed Bivalve Mollusk; Frozen Chum Salmon(Round,Dressed,Semi-Dressed,Fillet,Head,Bone,Skin); Frozen 1-35-1 Alaska Pollack(Round,Dressed,Semi-Dressed,Fillet); Frozen Pacific Taiyo Sangyo Co.,Ltd. 3 VN01840003 Showachuo,Kushiro- Cod(Round,Dressed,Semi-Dressed,Fillet); Frozen Pacific Saury(Round,Dressed,Semi- Kushiro Factory City,Hokkaido,Japan Dressed); Frozen Chub Mackerel(Round,Fillet); Frozen Blue Mackerel(Round,Fillet); Frozen Salted Pollack Roe 3-9 Komaba- Taiyo Sangyo Co.,Ltd. 4 VN01860004 Cho,Nemuro- Frozen Fish ; Frozen Processed Fish; (Excluding By-Product) Nemuro Factory City,Hokkaido,Japan 3-2-20 Kitahama- Marutoku Abe Suisan 5 VN01920005 Cho,Monbetu- Frozen Chum Salmon Dressed; Frozen Salmon Dressed Co.,Ltd City,Hokkaido,Japan Frozen Chum Salmon(Round,Semi-Dressed,Fillet); Frozen Salmon Milt; Frozen Pink Salmon(Round,Semi-Dressed,Dressed,Fillet); -
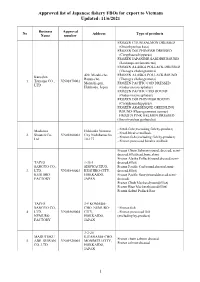
Approved List of Japanese Fishery Fbos for Export to Vietnam Updated: 11/6/2021
Approved list of Japanese fishery FBOs for export to Vietnam Updated: 11/6/2021 Business Approval No Address Type of products Name number FROZEN CHUM SALMON DRESSED (Oncorhynchus keta) FROZEN DOLPHINFISH DRESSED (Coryphaena hippurus) FROZEN JAPANESE SARDINE ROUND (Sardinops melanostictus) FROZEN ALASKA POLLACK DRESSED (Theragra chalcogramma) 420, Misaki-cho, FROZEN ALASKA POLLACK ROUND Kaneshin Rausu-cho, (Theragra chalcogramma) 1. Tsuyama CO., VN01870001 Menashi-gun, FROZEN PACIFIC COD DRESSED LTD Hokkaido, Japan (Gadus macrocephalus) FROZEN PACIFIC COD ROUND (Gadus macrocephalus) FROZEN DOLPHIN FISH ROUND (Coryphaena hippurus) FROZEN ARABESQUE GREENLING ROUND (Pleurogrammus azonus) FROZEN PINK SALMON DRESSED (Oncorhynchus gorbuscha) - Fresh fish (excluding fish by-product) Maekawa Hokkaido Nemuro - Fresh bivalve mollusk. 2. Shouten Co., VN01860002 City Nishihamacho - Frozen fish (excluding fish by-product) Ltd 10-177 - Frozen processed bivalve mollusk Frozen Chum Salmon (round, dressed, semi- dressed,fillet,head,bone,skin) Frozen Alaska Pollack(round,dressed,semi- TAIYO 1-35-1 dressed,fillet) SANGYO CO., SHOWACHUO, Frozen Pacific Cod(round,dressed,semi- 3. LTD. VN01840003 KUSHIRO-CITY, dressed,fillet) KUSHIRO HOKKAIDO, Frozen Pacific Saury(round,dressed,semi- FACTORY JAPAN dressed) Frozen Chub Mackerel(round,fillet) Frozen Blue Mackerel(round,fillet) Frozen Salted Pollack Roe TAIYO 3-9 KOMABA- SANGYO CO., CHO, NEMURO- - Frozen fish 4. LTD. VN01860004 CITY, - Frozen processed fish NEMURO HOKKAIDO, (excluding by-product) FACTORY JAPAN -

CALIFORNIA FISH and GAME "CONSERVATION of WILDLIFE THROUGH EDUCATION"
REPRINT FROM CALIFORNIA FISH and GAME "CONSERVATION OF WILDLIFE THROUGH EDUCATION" VOLUME 49 OCTOBER 1963 NUMBER 4 AN ORIENTAL GOBY COLLECTED IN THE SAN JOAQUIN RIVER DELTA NEAR STOCKTON, CALIFORNIA The California Department of Fish and Game has recently collected two specimens of a goby previously unknown in Californian waters. These we have identified as a species common in Japan, China, and Korea, Acanthogobius flavimanus ( Temminck and Schlegel), often re- ferred to as A. flavimanus (Schlegel). The first specimen was taken on January 18, 1963, by Arnold B. Albrecht and Vincent A. Catania (personnel of the Dingell-Johnson Project California F9R, "A Study of Sturgeon and Striped Bass," supported by Federal Aid to Fish Restoration funds), in a midwater trawl in the San Joaquin River off Prisoners Point on the southwest shore of Venice Island. Salinity here is very low, under 30 ppm (chlorides) at this time of year. The second specimen was collected in an otter trawl on March 29, 1963, by E. G. Gunderson, Armand P. Croft, Jr., and Vincent A. Catania, in the Stockton Deepwater Channel just above the entrance of the Calaveras River and just downstream from Port of Stockton. Although approxi- mately 12 miles above the first station, salinity here is slightly greater, measuring about 70 ppm (chlorides) at this period in the year. The first goby was 123 mm standard length (155 mm total length) ; the second was 69 mm s 1 (85 mm t.1.). Both have been deposited at Sacramento State College (SSC nos. 35-1 & 36-1). They exhibit close agreement with examples of A. -

National Noxious Fish List (Noxious in All Jurisdictions)
National noxious fish list (noxious in all jurisdictions) Family Specific name Common name Acestrorhynchidae Acestrorhynchus microlepis Alestiidae Hydrocynus spp Pike characin Giant tigerfish Amiidae Amia calva Bowfin Anabantidae Anabas testudineus Climbing perch Bagridae Anaspidoglanis macrostoma Flatnose catfish Bagrus ubangensis Ubangi shovelnose catfish Banded or spotted sunfish, largemouth bass, Centrarchidae — entire family bluegill Centropomidae Centropomus (12 spp) Snooks Lates microlepis Forktail lates Lates niloticus Nile perch Channidae Channa spp Snake head Chacidae Chaca chaca Angler, frogmouth and squarehead catfishes Characidae Colossoma spp Serrasalmus spp Redeye piranha Pygocentrus spp Red piranha Giant cichlid, yellow Cichlidae Boulengerochromis microlepis belly cichlid Oreochromis spp Tilapia Hemichromis fasciatus Banded jewelfish Pink, slender, greenwoods, mortimers,cunean and Sargochromis spp green happy Sarotherodon spp Sarotherodon melanotheron Blackchin tilapia Serranochromis spp Tilapia spp.(All except T. buttikoferi) Redbelly tilapia African pike-characin, tubenose poacher, fin Citharinidae entire subfamily Ichthyborinae eater Clariidae Clarias spp Walking catfish Cobitidae Misgurnus anguillicaudatus Weatherloach Cyprinidae Hypophthalmichthys nobilis Bighead carp Neolissochilus hexagonolepis Copper mahseer Gibelion catla Catla Catlocarpio siamensis Giant barb Cirrhinus cirrhosus Mrigal Ctenopharyngodon idella Grass carp Cyprinus carpio ‘European’ carp Labeo calbasu and L. rohita Orange fin labeo, rohu. Zacco platypus -

61661147.Pdf
Resource Inventory of Marine and Estuarine Fishes of the West Coast and Alaska: A Checklist of North Pacific and Arctic Ocean Species from Baja California to the Alaska–Yukon Border OCS Study MMS 2005-030 and USGS/NBII 2005-001 Project Cooperation This research addressed an information need identified Milton S. Love by the USGS Western Fisheries Research Center and the Marine Science Institute University of California, Santa Barbara to the Department University of California of the Interior’s Minerals Management Service, Pacific Santa Barbara, CA 93106 OCS Region, Camarillo, California. The resource inventory [email protected] information was further supported by the USGS’s National www.id.ucsb.edu/lovelab Biological Information Infrastructure as part of its ongoing aquatic GAP project in Puget Sound, Washington. Catherine W. Mecklenburg T. Anthony Mecklenburg Report Availability Pt. Stephens Research Available for viewing and in PDF at: P. O. Box 210307 http://wfrc.usgs.gov Auke Bay, AK 99821 http://far.nbii.gov [email protected] http://www.id.ucsb.edu/lovelab Lyman K. Thorsteinson Printed copies available from: Western Fisheries Research Center Milton Love U. S. Geological Survey Marine Science Institute 6505 NE 65th St. University of California, Santa Barbara Seattle, WA 98115 Santa Barbara, CA 93106 [email protected] (805) 893-2935 June 2005 Lyman Thorsteinson Western Fisheries Research Center Much of the research was performed under a coopera- U. S. Geological Survey tive agreement between the USGS’s Western Fisheries -

Report on the Monitoring of Radionuclides in Fishery Products (March 2011 – March 2016)
Report on the Monitoring of Radionuclides in Fishery Products (March 2011 – March 2016) October 2017 Fisheries Agency of Japan Table of Contents Overview ...................................................................................................................................... 8 The Purpose of this Report ............................................................................................................9 Part One. Efforts to Guarantee the Safety of Fishery Products ....................................................... 11 Chapter 1. Monitoring of Radioactive Materials in Food; Restrictions on Distribution and Other Countermeasures ..................................................................................................... 11 1-1-1 Standard Limits for Radioactive Materials in Food ............................................................... 11 1-1-2 Methods of Testing for Radioactive Materials ...................................................................... 12 1-1-3 Inspections of Fishery Products for Radioactive Materials ..................................................... 14 1-1-4 Restrictions and Suspensions on Distribution and Shipping ................................................... 17 1-1-5 Cancellation of Restrictions on Shipping and Distribution ..................................................... 19 Box 1 Calculation of the Limits for Human Consumption .............................................................. 22 Box 2 Survey of Radiation Dose from Radionuclides in Foods Calculation -

Mtdna Singletons As Evidence of a Post-Invasion Genetic Bottleneck in Yellowfin Goby Acanthogobius Flavimanus from San Francisco Bay, California
MARINE ECOLOGY PROGRESS SERIES Vol. 296: 197–208, 2005 Published July 12 Mar Ecol Prog Ser mtDNA singletons as evidence of a post-invasion genetic bottleneck in yellowfin goby Acanthogobius flavimanus from San Francisco Bay, California Matthew E. Neilson1, 2, Raymond R. Wilson Jr1,* 1Department of Biological Sciences, California State University, Long Beach, 1250 Bellflower Boulevard, Long Beach, California 90840, USA 2Present address: Great Lakes Genetics Lab, Lake Erie Center, University of Toledo, 6200 Bayshore Road, Oregon, Ohio 42618, USA ABSTRACT: Yellowfin goby, a fish native to East Asia, was first reported in northern California (San Francisco Bay) in 1963 and in southern California (Los Angeles Harbor) in 1979. Over the past 4 decades, it has spread to other northern and southern California estuaries, respectively. The docu- mented expansion of yellowfin goby in California provided an opportunity to study potential founder effects in a marine invasive fish by seeking evidence of reductions in mtDNA singleton haplotypes consistent with an expected loss of overall genetic diversity. We obtained samples of yellowfin goby from San Francisco Bay in northern California, from 2 small estuaries in southern California, and from Tokyo Bay, Japan, the presumed source population. The mtDNA control region was fully sequenced and analyzed for a total of 216 specimens, where the numbers of singleton haplotypes relative to the total number of haplotypes in each sample were compared to predictions from a regression analysis of singletons on total haplotypes. AMOVA comparisons among haplotype frequencies were also per- formed. Singleton haplotypes were significantly fewer than predicted among yellowfin goby of San Francisco Bay, indicating some loss of genetic diversity in that invasive population. -

The Marine Fish Parasite Nerocila Japonica (Isopoda: Cymothoidae
RESEARCH ARTICLES Nature of Kagoshima Vol. 46 The marine fish parasite Nerocila japonica (Isopoda: Cymothoidae) from a euryhaline cyprinid fish (Pseudaspius hakonensis) in brackish waters of a river in central Japan Kazuya Nagasawa1,2 and Nobuo Inoue3 1Graduate School of Integrated Sciences for Life, Hiroshima University, 1–4–4 Kagamiyama, Higashi-Hiroshima, Hiroshima 739–8528, Japan 2Aquaparasitology Laboratory, 365–61 Kusanagi, Shizuoka 424–0886, Japan 3Biodiversity Network Niigata, 1–8–25 Terayama, Higashi, Niigata 950–0892, Japan ■ Abstract Island, Niigata Prefecture, central Japan. This is inter- A juvenile of Nerocila japonica Schioedte and Mein- esting because N. japonica is a marine fish parasite but ert, 1881 was found parasitizing the ventral fin of a big- was collected in the river. The present paper reports on scaled redfin, Pseudaspius hakonensis (Günther, 1877) this collection and discusses the occurrence of N. ja- (Cypriniformes: Cyprinidae), in brackish waters of the ponica on euryhaline fishes in brackish waters. Ten-no River on Sado Island, Niigata Prefecture, cen- tral Japan. The river flows into brackish Lake Kamo, ■ Materials and Methods which is connected to the southern Sea of Japan. The The big-scaled redfin was caught using a small setnet big-scaled redfin is an unusual cyprinid that can inhabit on 8 November 2012 in the lower reaches of the Ten-no marine and brackish waters as well as fresh waters, and River at Niibo-katagami, Sado City, Niigata Prefecture, the fish caught most probably became infected by N. central Japan. The river, originating from the nearby the japonica at sea or in the lake and moved upstream in hill area, is small (1–5 m wide, 5 km in total length) and the river.