Cross-Species Evaluation of Molecular Target Sequence and Structural
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Article Evolutionary Dynamics of the OR Gene Repertoire in Teleost Fishes
bioRxiv preprint doi: https://doi.org/10.1101/2021.03.09.434524; this version posted March 10, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Article Evolutionary dynamics of the OR gene repertoire in teleost fishes: evidence of an association with changes in olfactory epithelium shape Maxime Policarpo1, Katherine E Bemis2, James C Tyler3, Cushla J Metcalfe4, Patrick Laurenti5, Jean-Christophe Sandoz1, Sylvie Rétaux6 and Didier Casane*,1,7 1 Université Paris-Saclay, CNRS, IRD, UMR Évolution, Génomes, Comportement et Écologie, 91198, Gif-sur-Yvette, France. 2 NOAA National Systematics Laboratory, National Museum of Natural History, Smithsonian Institution, Washington, D.C. 20560, U.S.A. 3Department of Paleobiology, National Museum of Natural History, Smithsonian Institution, Washington, D.C., 20560, U.S.A. 4 Independent Researcher, PO Box 21, Nambour QLD 4560, Australia. 5 Université de Paris, Laboratoire Interdisciplinaire des Energies de Demain, Paris, France 6 Université Paris-Saclay, CNRS, Institut des Neurosciences Paris-Saclay, 91190, Gif-sur- Yvette, France. 7 Université de Paris, UFR Sciences du Vivant, F-75013 Paris, France. * Corresponding author: e-mail: [email protected]. !1 bioRxiv preprint doi: https://doi.org/10.1101/2021.03.09.434524; this version posted March 10, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Teleost fishes perceive their environment through a range of sensory modalities, among which olfaction often plays an important role. -

§4-71-6.5 LIST of CONDITIONALLY APPROVED ANIMALS November
§4-71-6.5 LIST OF CONDITIONALLY APPROVED ANIMALS November 28, 2006 SCIENTIFIC NAME COMMON NAME INVERTEBRATES PHYLUM Annelida CLASS Oligochaeta ORDER Plesiopora FAMILY Tubificidae Tubifex (all species in genus) worm, tubifex PHYLUM Arthropoda CLASS Crustacea ORDER Anostraca FAMILY Artemiidae Artemia (all species in genus) shrimp, brine ORDER Cladocera FAMILY Daphnidae Daphnia (all species in genus) flea, water ORDER Decapoda FAMILY Atelecyclidae Erimacrus isenbeckii crab, horsehair FAMILY Cancridae Cancer antennarius crab, California rock Cancer anthonyi crab, yellowstone Cancer borealis crab, Jonah Cancer magister crab, dungeness Cancer productus crab, rock (red) FAMILY Geryonidae Geryon affinis crab, golden FAMILY Lithodidae Paralithodes camtschatica crab, Alaskan king FAMILY Majidae Chionocetes bairdi crab, snow Chionocetes opilio crab, snow 1 CONDITIONAL ANIMAL LIST §4-71-6.5 SCIENTIFIC NAME COMMON NAME Chionocetes tanneri crab, snow FAMILY Nephropidae Homarus (all species in genus) lobster, true FAMILY Palaemonidae Macrobrachium lar shrimp, freshwater Macrobrachium rosenbergi prawn, giant long-legged FAMILY Palinuridae Jasus (all species in genus) crayfish, saltwater; lobster Panulirus argus lobster, Atlantic spiny Panulirus longipes femoristriga crayfish, saltwater Panulirus pencillatus lobster, spiny FAMILY Portunidae Callinectes sapidus crab, blue Scylla serrata crab, Samoan; serrate, swimming FAMILY Raninidae Ranina ranina crab, spanner; red frog, Hawaiian CLASS Insecta ORDER Coleoptera FAMILY Tenebrionidae Tenebrio molitor mealworm, -

Ecography ECOG-01937 Hattab, T., Leprieur, F., Ben Rais Lasram, F., Gravel, D., Le Loc’H, F
Ecography ECOG-01937 Hattab, T., Leprieur, F., Ben Rais Lasram, F., Gravel, D., Le Loc’h, F. and Albouy, C. 2016. Forecasting fine- scale changes in the food-web structure of coastal marine communities under climate change. – Ecography doi: 10.1111/ecog.01937 Supplementary material Forecasting fine-scale changes in the food-web structure of coastal marine communities under climate change by Hattab et al. Appendix 1 List of coastal exploited marine species considered in this study Species Genus Order Family Class Trophic guild Auxis rochei rochei (Risso, 1810) Auxis Perciformes Scombridae Actinopterygii Top predators Balistes capriscus Gmelin, 1789 Balistes Tetraodontiformes Balistidae Actinopterygii Macro-carnivorous Boops boops (Linnaeus, 1758) Boops Perciformes Sparidae Actinopterygii Basal species Carcharhinus plumbeus (Nardo, 1827) Carcharhinus Carcharhiniformes Carcharhinidae Elasmobranchii Top predators Dasyatis pastinaca (Linnaeus, 1758) Dasyatis Rajiformes Dasyatidae Elasmobranchii Top predators Dentex dentex (Linnaeus, 1758) Dentex Perciformes Sparidae Actinopterygii Macro-carnivorous Dentex maroccanus Valenciennes, 1830 Dentex Perciformes Sparidae Actinopterygii Macro-carnivorous Diplodus annularis (Linnaeus, 1758) Diplodus Perciformes Sparidae Actinopterygii Forage species Diplodus sargus sargus (Linnaeus, 1758) Diplodus Perciformes Sparidae Actinopterygii Macro-carnivorous (Geoffroy Saint- Diplodus vulgaris Hilaire, 1817) Diplodus Perciformes Sparidae Actinopterygii Basal species Engraulis encrasicolus (Linnaeus, 1758) Engraulis -

A Practical Handbook for Determining the Ages of Gulf of Mexico And
A Practical Handbook for Determining the Ages of Gulf of Mexico and Atlantic Coast Fishes THIRD EDITION GSMFC No. 300 NOVEMBER 2020 i Gulf States Marine Fisheries Commission Commissioners and Proxies ALABAMA Senator R.L. “Bret” Allain, II Chris Blankenship, Commissioner State Senator District 21 Alabama Department of Conservation Franklin, Louisiana and Natural Resources John Roussel Montgomery, Alabama Zachary, Louisiana Representative Chris Pringle Mobile, Alabama MISSISSIPPI Chris Nelson Joe Spraggins, Executive Director Bon Secour Fisheries, Inc. Mississippi Department of Marine Bon Secour, Alabama Resources Biloxi, Mississippi FLORIDA Read Hendon Eric Sutton, Executive Director USM/Gulf Coast Research Laboratory Florida Fish and Wildlife Ocean Springs, Mississippi Conservation Commission Tallahassee, Florida TEXAS Representative Jay Trumbull Carter Smith, Executive Director Tallahassee, Florida Texas Parks and Wildlife Department Austin, Texas LOUISIANA Doug Boyd Jack Montoucet, Secretary Boerne, Texas Louisiana Department of Wildlife and Fisheries Baton Rouge, Louisiana GSMFC Staff ASMFC Staff Mr. David M. Donaldson Mr. Bob Beal Executive Director Executive Director Mr. Steven J. VanderKooy Mr. Jeffrey Kipp IJF Program Coordinator Stock Assessment Scientist Ms. Debora McIntyre Dr. Kristen Anstead IJF Staff Assistant Fisheries Scientist ii A Practical Handbook for Determining the Ages of Gulf of Mexico and Atlantic Coast Fishes Third Edition Edited by Steve VanderKooy Jessica Carroll Scott Elzey Jessica Gilmore Jeffrey Kipp Gulf States Marine Fisheries Commission 2404 Government St Ocean Springs, MS 39564 and Atlantic States Marine Fisheries Commission 1050 N. Highland Street Suite 200 A-N Arlington, VA 22201 Publication Number 300 November 2020 A publication of the Gulf States Marine Fisheries Commission pursuant to National Oceanic and Atmospheric Administration Award Number NA15NMF4070076 and NA15NMF4720399. -

Tanichthys Kuehnei, New Species, from Central Vietnam (Cypriniformes: Cyprinidae)
1 Ichthyological Exploration of Freshwaters/IEF-1081/pp. 1-10 Published 9 February 2019 LSID: http://zoobank.org/urn:lsid:zoobank.org:pub:7F247BBC-6CC9-447A-8F88-ECACA2CB0934 DOI: http://doi.org/10.23788/IEF-1081 Tanichthys kuehnei, new species, from Central Vietnam (Cypriniformes: Cyprinidae) Jörg Bohlen*, Tomáš Dvorák*, **, Ha Nam Thang*** and Vendula Šlechtová* Tanichthys kuehnei, new species, is described from a stream in the Bach Ma Mountains in Hue Province in Central 1 1 Vietnam. The new species differs from its congeners by having more branched rays in anal fin (9 /2 vs. 7-8 /2 1 in T. micagemmae and 8 /2 in T. albonubes and T. thacbaensis). Morphological and genetic characters suggest it to be closer related to T. micagemmae, the only other species of Tanichthys known from Central Vietnam. Tanichthys kuehnei differs from T. micagemmae by having a white anal-fin margin (vs. red). Introduction genus Tanichthys inhabit moderately large to very small streams, with populations frequently being Cyprinid fishes of the genus Tanichthys are small restricted to very small geographic areas and are (maximum 33 mm SL) but colourful and well- often found only very locally (Freyhof & Herder, liked by ornamental fish hobbyists. The genus is 2001), leading to strong isolation effects between characterised by confluent narial openings that are the populations. This isolation was demonstrated not separated by a skin wall and by males bear- in a genetic study by Luo et al. (2015) where each ing cornified tubercles on the snout posterior to of six analysed wild populations of T. albonubes premaxilla (Freyhof & Herder, 2001). -

United States National Museum Bulletin 282
Cl>lAat;i<,<:>';i^;}Oit3Cl <a f^.S^ iVi^ 5' i ''*«0£Mi»«33'**^ SMITHSONIAN INSTITUTION MUSEUM O F NATURAL HISTORY I NotUTus albater, new species, a female paratype, 63 mm. in standard length; UMMZ 102781, Missouri. (Courtesy Museum of Zoology, University of Michigan.) UNITED STATES NATIONAL MUSEUM BULLETIN 282 A Revision of the Catfish Genus Noturus Rafinesque^ With an Analysis of Higher Groups in the Ictaluridae WILLIAM RALPH TAYLOR Associate Curator, Division of Fishes SMITHSONIAN INSTITUTION PRESS CITY OF WASHINGTON 1969 IV Publications of the United States National Museum The scientific publications of the United States National Museum include two series, Proceedings of the United States National Museum and United States National Museum Bulletin. In these series are published original articles and monographs dealing with the collections and work of the Museum and setting forth newly acquired facts in the fields of anthropology, biology, geology, history, and technology. Copies of each publication are distributed to libraries and scientific organizations and to specialists and others interested in the various subjects. The Proceedings, begun in 1878, are intended for the publication, in separate form, of shorter papers. These are gathered in volumes, octavo in size, with the publication date of each paper recorded in the table of contents of the volume. In the Bulletin series, the first of which was issued in 1875, appear longer, separate publications consisting of monographs (occasionally in several parts) and volumes in which are collected works on related subjects. Bulletins are either octavo or quarto in size, depending on the needs of the presentation. Since 1902, papers relating to the botanical collections of the Museum have been published in the Bulletin series under the heading Contributions from the United States National Herbarium. -

Fish, Crustaceans, Molluscs, Etc Capture Production by Species
440 Fish, crustaceans, molluscs, etc Capture production by species items Asia - Inland waters C-04 Poissons, crustacés, mollusques, etc Captures par catégories d'espèces Asie - Eaux continentales (a) Peces, crustáceos, moluscos, etc Capturas por categorías de especies Asia - Aguas continentales English name Scientific name Species group Nom anglais Nom scientifique Groupe d'espèces 2003 2004 2005 2006 2007 2008 2009 Nombre inglés Nombre científico Grupo de especies t t t t t t t Freshwater bream Abramis brama 11 16 375 22 345 21 073 17 834 19 313 22 083 17 612 Freshwater breams nei Abramis spp 11 33 27 92 13 11 9 1 Common carp Cyprinus carpio 11 36 617 39 126 39 104 32 466 34 723 46 360 42 166 Tench Tinca tinca 11 935 2 082 1 792 1 953 1 884 1 632 1 482 Crucian carp Carassius carassius 11 4 330 3 538 4 621 3 429 3 865 4 921 3 656 Goldfish Carassius auratus 11 1 1 2 2 3 1 1 Roach Rutilus rutilus 11 149 287 358 420 521 577 535 Kutum Rutilus frisii 11 9 015 7 052 9 649 16 138 17 242 14 885 12 536 Roaches nei Rutilus spp 11 1 450 1 700 1 537 2 088 2 383 4 848 4 211 Common dace Leuciscus leuciscus 11 82 93 87 85 82 71 63 Mud carp Cirrhinus molitorella 11 11 10 8 - - - - Grass carp(=White amur) Ctenopharyngodon idellus 11 1 959 2 458 3 066 3 312 4 417 4 215 4 567 Beardless barb Cyclocheilichthys apogon 11 .. -
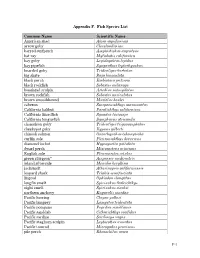
Appendix E: Fish Species List
Appendix F. Fish Species List Common Name Scientific Name American shad Alosa sapidissima arrow goby Clevelandia ios barred surfperch Amphistichus argenteus bat ray Myliobatis californica bay goby Lepidogobius lepidus bay pipefish Syngnathus leptorhynchus bearded goby Tridentiger barbatus big skate Raja binoculata black perch Embiotoca jacksoni black rockfish Sebastes melanops bonehead sculpin Artedius notospilotus brown rockfish Sebastes auriculatus brown smoothhound Mustelus henlei cabezon Scorpaenichthys marmoratus California halibut Paralichthys californicus California lizardfish Synodus lucioceps California tonguefish Symphurus atricauda chameleon goby Tridentiger trigonocephalus cheekspot goby Ilypnus gilberti chinook salmon Oncorhynchus tshawytscha curlfin sole Pleuronichthys decurrens diamond turbot Hypsopsetta guttulata dwarf perch Micrometrus minimus English sole Pleuronectes vetulus green sturgeon* Acipenser medirostris inland silverside Menidia beryllina jacksmelt Atherinopsis californiensis leopard shark Triakis semifasciata lingcod Ophiodon elongatus longfin smelt Spirinchus thaleichthys night smelt Spirinchus starksi northern anchovy Engraulis mordax Pacific herring Clupea pallasi Pacific lamprey Lampetra tridentata Pacific pompano Peprilus simillimus Pacific sanddab Citharichthys sordidus Pacific sardine Sardinops sagax Pacific staghorn sculpin Leptocottus armatus Pacific tomcod Microgadus proximus pile perch Rhacochilus vacca F-1 plainfin midshipman Porichthys notatus rainwater killifish Lucania parva river lamprey Lampetra -

Report on the Monitoring of Radionuclides in Fishery Products (March 2011 - January 2015)
Report on the Monitoring of Radionuclides in Fishery Products (March 2011 - January 2015) April 2015 Fisheries Agency of Japan 0 1 Table of Contents Overview…………………………………………………………………………………………………. 8 The Purpose of this Report………………………………………………………………………………9 Part One. Efforts to Guarantee the Safety of Fishery Products………………………………………..11 Chapter 1. Monitoring of Radioactive Materials in Food; Restrictions on Distribution and Other Countermeasures………...…………………………………………………………………11 1-1-1 Standard Limits for Radioactive Materials in Food………………………………………...……11 1-1-2 Methods of Testing for Radioactive Materials………………………………………...…………12 1-1-3 Inspections of Fishery Products for Radioactive Materials…………………………...…………14 1-1-4 Restrictions and Suspensions on Distribution and Shipping ……………………………………..18 1-1-5 Cancellation of Restrictions on Shipping and Distribution………………………………………20 Box 1 Calculation of the Limits for Human Consumption……..………………………………………23 Box 2 Survey of Radiation Dose from Radionuclides in Foods Calculation of the Limits…………….24 Box 3 Examples of Local Government Monitoring Plan………………………………...…………….25 Chapter 2. Results of Radioactive Cesium Inspections for Fishery Products…………………………26 1-2-1 Inspection Results for Nationwide Fishery Products in Japan (in total)…………………………26 1-2-2 Inspection Results for Fukushima Prefecture Fishery Products (all)…………………………….27 1-2-3 Inspection Results for Fishery Products (all) from Outside Fukushima Prefecture……………...30 1-2-4 Trends within Fish Species……………………………………………………………………….32 1-2-5 Inspection Results for Main Target Fish Species of Fishing and Farming by Fiscal Year……….42 1-2-6 Radioactive Material Concentrations within Fish within 20 km of the Fukushima Daiichi NPS.46 Box 4 Fukushima Fishing Trials………………………………...……………………………………...47 1-2-7 Screening Test by Prefectural and Municipal Governments……………………………………..48 Chapter 3. Inspection for Radionuclides Other Than Radioactive Cesium……………………………49 1-3-1 Inspections for Radioactive Strontium etc. -

Composition and Abundance of Drifting Fish Eggs on the Upper Reaches of Xijiang
bioRxiv preprint doi: https://doi.org/10.1101/2020.01.13.904110; this version posted January 13, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 Composition and Abundance of Drifting Fish Eggs on the Upper Reaches of Xijiang 2 River, China, after the Formation of the Cascade Reservoirs 3 4 Short title: Composition and Abundance of Drifting Fish Eggs in the Upper Reaches of a 5 River after Dam Formation 6 Gao Minghui1, Wu Zhiqiang2, 3, Tan Xichang4, Huang Liangliang3, Huang Haibo3 and Liu Hao3 7 8 1 College of Life Science and Technology, Guangxi University, Nanning 350000, Guangxi, 9 China 10 2 School of Marine Sciences, Guangxi University, Nanning 350000, Guangxi, China 11 3 College of Environmental Science and Engineering, Guilin University of Technology, Guilin 12 541000, Guangxi, China. 13 4 ZhuJiang Water Conservancy Bureau, Ministry of Water Resources, Guangzhou 510000, 14 Guangdong, China 15 16 Corresponding Author: 17 Wu Zhiqiang 18 Yanshan Town, Guilin, Guangxi 54100, China 19 Email address: [email protected] 20 21 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.01.13.904110; this version posted January 13, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. -

A Dissertation Entitled Evolution, Systematics
A Dissertation Entitled Evolution, systematics, and phylogeography of Ponto-Caspian gobies (Benthophilinae: Gobiidae: Teleostei) By Matthew E. Neilson Submitted as partial fulfillment of the requirements for The Doctor of Philosophy Degree in Biology (Ecology) ____________________________________ Adviser: Dr. Carol A. Stepien ____________________________________ Committee Member: Dr. Christine M. Mayer ____________________________________ Committee Member: Dr. Elliot J. Tramer ____________________________________ Committee Member: Dr. David J. Jude ____________________________________ Committee Member: Dr. Juan L. Bouzat ____________________________________ College of Graduate Studies The University of Toledo December 2009 Copyright © 2009 This document is copyrighted material. Under copyright law, no parts of this document may be reproduced without the expressed permission of the author. _______________________________________________________________________ An Abstract of Evolution, systematics, and phylogeography of Ponto-Caspian gobies (Benthophilinae: Gobiidae: Teleostei) Matthew E. Neilson Submitted as partial fulfillment of the requirements for The Doctor of Philosophy Degree in Biology (Ecology) The University of Toledo December 2009 The study of biodiversity, at multiple hierarchical levels, provides insight into the evolutionary history of taxa and provides a framework for understanding patterns in ecology. This is especially poignant in invasion biology, where the prevalence of invasiveness in certain taxonomic groups could -

Summary Report of Freshwater Nonindigenous Aquatic Species in U.S
Summary Report of Freshwater Nonindigenous Aquatic Species in U.S. Fish and Wildlife Service Region 4—An Update April 2013 Prepared by: Pam L. Fuller, Amy J. Benson, and Matthew J. Cannister U.S. Geological Survey Southeast Ecological Science Center Gainesville, Florida Prepared for: U.S. Fish and Wildlife Service Southeast Region Atlanta, Georgia Cover Photos: Silver Carp, Hypophthalmichthys molitrix – Auburn University Giant Applesnail, Pomacea maculata – David Knott Straightedge Crayfish, Procambarus hayi – U.S. Forest Service i Table of Contents Table of Contents ...................................................................................................................................... ii List of Figures ............................................................................................................................................ v List of Tables ............................................................................................................................................ vi INTRODUCTION ............................................................................................................................................. 1 Overview of Region 4 Introductions Since 2000 ....................................................................................... 1 Format of Species Accounts ...................................................................................................................... 2 Explanation of Maps ................................................................................................................................