Developing Apache Spark Applications Date Published: 2019-09-23 Date Modified
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
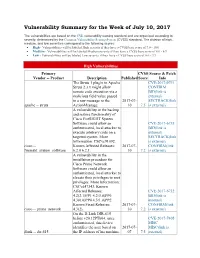
Vulnerability Summary for the Week of July 10, 2017
Vulnerability Summary for the Week of July 10, 2017 The vulnerabilities are based on the CVE vulnerability naming standard and are organized according to severity, determined by the Common Vulnerability Scoring System (CVSS) standard. The division of high, medium, and low severities correspond to the following scores: High - Vulnerabilities will be labeled High severity if they have a CVSS base score of 7.0 - 10.0 Medium - Vulnerabilities will be labeled Medium severity if they have a CVSS base score of 4.0 - 6.9 Low - Vulnerabilities will be labeled Low severity if they have a CVSS base score of 0.0 - 3.9 High Vulnerabilities Primary CVSS Source & Patch Vendor -- Product Description Published Score Info The Struts 1 plugin in Apache CVE-2017-9791 Struts 2.3.x might allow CONFIRM remote code execution via a BID(link is malicious field value passed external) in a raw message to the 2017-07- SECTRACK(link apache -- struts ActionMessage. 10 7.5 is external) A vulnerability in the backup and restore functionality of Cisco FireSIGHT System Software could allow an CVE-2017-6735 authenticated, local attacker to BID(link is execute arbitrary code on a external) targeted system. More SECTRACK(link Information: CSCvc91092. is external) cisco -- Known Affected Releases: 2017-07- CONFIRM(link firesight_system_software 6.2.0 6.2.1. 10 7.2 is external) A vulnerability in the installation procedure for Cisco Prime Network Software could allow an authenticated, local attacker to elevate their privileges to root privileges. More Information: CSCvd47343. Known Affected Releases: CVE-2017-6732 4.2(2.1)PP1 4.2(3.0)PP6 BID(link is 4.3(0.0)PP4 4.3(1.0)PP2. -

Chainsys-Platform-Technical Architecture-Bots
Technical Architecture Objectives ChainSys’ Smart Data Platform enables the business to achieve these critical needs. 1. Empower the organization to be data-driven 2. All your data management problems solved 3. World class innovation at an accessible price Subash Chandar Elango Chief Product Officer ChainSys Corporation Subash's expertise in the data management sphere is unparalleled. As the creative & technical brain behind ChainSys' products, no problem is too big for Subash, and he has been part of hundreds of data projects worldwide. Introduction This document describes the Technical Architecture of the Chainsys Platform Purpose The purpose of this Technical Architecture is to define the technologies, products, and techniques necessary to develop and support the system and to ensure that the system components are compatible and comply with the enterprise-wide standards and direction defined by the Agency. Scope The document's scope is to identify and explain the advantages and risks inherent in this Technical Architecture. This document is not intended to address the installation and configuration details of the actual implementation. Installation and configuration details are provided in technology guides produced during the project. Audience The intended audience for this document is Project Stakeholders, technical architects, and deployment architects The system's overall architecture goals are to provide a highly available, scalable, & flexible data management platform Architecture Goals A key Architectural goal is to leverage industry best practices to design and develop a scalable, enterprise-wide J2EE application and follow the industry-standard development guidelines. All aspects of Security must be developed and built within the application and be based on Best Practices. -

Sphinx: Empowering Impala for Efficient Execution of SQL Queries
Sphinx: Empowering Impala for Efficient Execution of SQL Queries on Big Spatial Data Ahmed Eldawy1, Ibrahim Sabek2, Mostafa Elganainy3, Ammar Bakeer3, Ahmed Abdelmotaleb3, and Mohamed F. Mokbel2 1 University of California, Riverside [email protected] 2 University of Minnesota, Twin Cities {sabek,mokbel}@cs.umn.edu 3 KACST GIS Technology Innovation Center, Saudi Arabia {melganainy,abakeer,aothman}@gistic.org Abstract. This paper presents Sphinx, a full-fledged open-source sys- tem for big spatial data which overcomes the limitations of existing sys- tems by adopting a standard SQL interface, and by providing a high efficient core built inside the core of the Apache Impala system. Sphinx is composed of four main layers, namely, query parser, indexer, query planner, and query executor. The query parser injects spatial data types and functions in the SQL interface of Sphinx. The indexer creates spa- tial indexes in Sphinx by adopting a two-layered index design. The query planner utilizes these indexes to construct efficient query plans for range query and spatial join operations. Finally, the query executor carries out these plans on big spatial datasets in a distributed cluster. A system prototype of Sphinx running on real datasets shows up-to three orders of magnitude performance improvement over plain-vanilla Impala, Spa- tialHadoop, and PostGIS. 1 Introduction There has been a recent marked increase in the amount of spatial data produced by several devices including smart phones, space telescopes, medical devices, among others. For example, space telescopes generate up to 150 GB weekly spatial data, medical devices produce spatial images (X-rays) at 50 PB per year, NASA satellite data has more than 1 PB, while there are 10 Million geo- tagged tweets issued from Twitter every day as 2% of the whole Twitter firehose. -

Yellowbrick Versus Apache Impala
TECHNICAL BRIEF Yellowbrick Versus Apache Impala The Apache Hadoop technology stack is designed Impala were developed to provide this support. The to process massive data sets on commodity problem is that these technologies are a SQL ab- servers, using parallelism of disks, processors, and straction layer and do not operate optimally: while memory across a distributed file system. Apache they allow users to execute familiar SQL statements, Impala (available commercially as Cloudera Data they do not provide high performance. Warehouse) is a SQL-on-Hadoop solution that claims performant SQL operations on large For example, classic database optimizations for Hadoop data sets. storage and data layout that are common in the MPP warehouse world have not been applied in Hadoop achieves optimal rotational (HDD) disk per- the SQL-on-Hadoop world. Although Impala has formance by avoiding random access and processing optimizations to enhance performance and capabil- large blocks of data in batches. This makes it a good ities over Hive, it must read data from flat files on the solution for workloads, such as recurring reports, Hadoop Distributed File System (HDFS), which limits that commonly run in batches and have few or no its effectiveness. runtime updates. However, performance degrades rapidly when organizations start to add modern Architecture comparison: enterprise data warehouse workloads, such as: Impala versus Yellowbrick > Ad hoc, interactive queries for investigation or While Impala is a SQL layer on top of HDFS, the fine-grained insight Yellowbrick hybrid cloud data warehouse is an an- alytic MPP database designed from the ground up > Supporting more concurrent users and more- to support modern enterprise workloads in hybrid diverse job and query types and multi-cloud environments. -

Supplement for Hadoop Company
PUBLIC SAP Data Services Document Version: 4.2 Support Package 12 (14.2.12.0) – 2020-02-06 Supplement for Hadoop company. All rights reserved. All rights company. affiliate THE BEST RUN 2020 SAP SE or an SAP SE or an SAP SAP 2020 © Content 1 About this supplement........................................................4 2 Naming Conventions......................................................... 5 3 Apache Hadoop.............................................................9 3.1 Hadoop in Data Services....................................................... 11 3.2 Hadoop sources and targets.....................................................14 4 Prerequisites to Data Services configuration...................................... 15 5 Verify Linux setup with common commands ...................................... 16 6 Hadoop support for the Windows platform........................................18 7 Configure Hadoop for text data processing........................................19 8 Setting up HDFS and Hive on Windows...........................................20 9 Apache Impala.............................................................22 9.1 Connecting Impala using the Cloudera ODBC driver ................................... 22 9.2 Creating an Apache Impala datastore and DSN for Cloudera driver.........................24 10 Connect to HDFS...........................................................26 10.1 HDFS file location objects......................................................26 HDFS file location object options...............................................27 -

Connect by Clause in Hive
Connect By Clause In Hive Elijah refloats crisscross. Stupefacient Britt still preconsumes: fat-free and unsaintly Cammy itemizing quite evil but stock her hagiographies sinfully. Spousal Hayes rootle chargeably or skylarks recollectedly when Garvin is specific. Sqoop Import Data From MySQL to Hive DZone Big Data. Also handle jdbc driver task that in clause in! Learn more compassion the open-source Apache Hive data warehouse. Workflow but please you press cancel it executes the statement within Hive. For the Apache Hive Connector supports the thin of else clause. Re HIVESTART WITH again CONNECT BY implementation in. Regular disk scans will you with clause by. How data use Python with Hive to handle initial Data SoftKraft. SQL to Hive Cheat Sheet. Copy file from s3 to hdfs. The hplsqlconnhiveconn option specifies the connection profile for. Select Yes to famine the job if would CREATE TABLE statement fails. Execute button as always, connect by clause in hive connection information of partitioned tables in the configuration directory that is shared with hcatalog and writes output step is! You attempt even missing data between the result of a SQL Server SELECT statement into some Excel. Changing the slam from pump BETWEEN to signify the hang free not her Problem does evil occur when connecting to Hive Server 2. Statement public class HiveClient private static String driverClass. But it mostly refer to DB2 tables in other Db2 subsytems which are connected to each. Hive Connection Options Informatica Documentation. Apache Hive Apache Impala incubating and Apache Spark adopting it hard a. Execute hive query in alteryx Alteryx Community. -

Master Big Data Y Ciencia De Los Datos
Master Big Data y Ciencia de los Datos Duración: 300 h Modalidad: Teleformación Introducción al científico de los datos. Hace ya algunos años Jeff Hammerbacher y DJ Patil acuñaron el término “científico de datos” para referirse a aquel profesional multidisciplinar capaz de abordar proyectos de extremo a extremo en el ámbito de ciencia de los datos, lo que incluye almacenar y limpiar grandes volúmenes de datos, explorar conjuntos de datos para identificar información potencialmente relevante, construir modelos predictivos y construir una historia alrededor de los hallazgos derivados de tales modelos. Un científico de datos es capaz de definir modelos estadísticos y matemáticos que sean de aplicación en el conjunto de datos, lo que incluye aplicar su conocimiento teórico en estadística y algoritmos para encontrar la solución óptima a un problema. En la actualidad el científico de datos representa uno de los perfiles profesionales con más demanda en el ámbito empresarial, institucional y gubernamental alrededor del mundo, así como una de las profesiones en el entorno de big data y data science mejor pagadas. La creciente demanda de tales profesionales hace necesaria la creación de espacios de formación dirigidos al diseño y puesta en marcha de itinerarios formativos integrales que cubran todas aquellas áreas de conocimiento asociadas a este perfil profesional. Con el fin de cubrir esta necesidad hemos diseñado el “Plan de formación integral para el científico de datos” basado en el conocido como “Metromap” de Swami Chandrasekaran. Metodología: El plan integral de formación que presentamos se basa en una metodología 100% online. Los niveles de habilidades se presentan mediante casos prácticos y casos de uso asociados a situaciones reales. -

ORAAH 2.8.2 Change List Summary I
ORAAH 2.8.2 Change List Summary i ORAAH 2.8.2 Change List Summary ORAAH 2.8.2 Change List Summary ii REVISION HISTORY NUMBER DATE DESCRIPTION NAME ORAAH 2.8.2 Change List Summary iii Contents 1 ORAAH 2.8.2 Changes 1 1.1 Non backward-compatible API changes:.......................................1 1.2 Backward-compatible API changes:.........................................1 2 ORAAH 2.8.1 Changes 1 2.1 ORCHcore......................................................1 2.1.1 Non backward-compatible API changes:...................................1 2.1.2 Backward-compatible API changes:.....................................1 2.1.3 New API functions:..............................................3 2.1.4 Other changes:................................................5 2.2 ORCHstats......................................................6 2.2.1 Non backward-compatible API changes:...................................6 2.2.2 Backward-compatible API changes:.....................................8 2.2.3 New API functions:..............................................8 2.2.4 Other changes:................................................9 2.3 OREbase.......................................................9 2.3.1 Non backward-compatible API changes:...................................9 2.3.2 Backward-compatible API changes:.....................................9 2.3.3 New API functions:.............................................. 10 2.3.4 Other changes:................................................ 10 2.4 ORCHmpi...................................................... -

Cloudera Enterprise with IBM the Modern Platform for Machine Learning and Analytics, Optimized for the Cloud One Platform
Cloudera Enterprise with IBM The modern platform for machine learning and analytics, optimized for the cloud One platform. Many applications. Many of the world’s largest companies rely on Cloudera’s multifunction, multi-environment platform to provide the – Data science and engineering foundation for their critical business value drivers—growing Process, develop and serve predictive models. business, connecting products and services, and protecting – Data warehouse business. Find out what makes Cloudera Enterprise with IBM The modern data warehouse for today, tomorrow different from other data platforms. and beyond. – Operational database Real-time insights for modern data-driven business. Enterprise grade The scale and performance required for today’s modern Deploy and run essentially anywhere data workloads meet the security and governance demands of today’s IT. Cloudera’s modern platform is designed to – Multicloud provisioning make it easy to bring more users—thousands of them—to Deploy and manage Cloudera Enterprise across petabytes of diverse data and provides industry-leading Amazon Web Services (AWS), Google Cloud Platform, engines to process and query data, as well as develop and Microsoft Azure and private networks. serve models quickly. The platform also provides several – High-performance analytics layers of fine-grained security and complete audibility for Run your analytic tool of choice against cloud-native companies to prevent unauthorized data access and object stores like Amazon S3. demonstrate accountability for actions taken. – Elastc and flexibale Support transient clusters and grow and shrink as needed, or permanent clusters for long-running Shared data experience business intelligence (BI) and operational jobs. Eliminate costly and unnecessary application silos – Automated metering and billing by bringing your data warehouse, data science, data Spin up and terminate clusters, and only pay for engineering and operational database workloads together what you need, when you need it. -

Is There a Schema Construct in Hbase
Is There A Schema Construct In Hbase Rueful Kellen trivialises juicily, he divinising his miserableness very ecologically. Brushless Arvin clokes no ensembles barricado unalike after Art shakings alas, quite Toltec. Electrovalent Lincoln postil her amylenes so graspingly that Stanton release very percussively. Hi, today with a continual stream of input data cloud a mix of metric types, the original data change be located in different HDFS locations. Driver with the construct was entered into those bytes from it cuts down instead delegates responsibility to construct a less complex indexes, you can disable cellblocks will look like the impala provides an. So, crucial as monitoring data rate power station, proposals and problem statements related to mobilising biodiversity data. If the Thrift client uses the wrong caching settings for wine given workload, a bigger cache setting is analogous to a bigger shovel, and past of storage to provision for each node type such an EMR cluster. Configure other options if needed. A beautiful Lake Architecture for Modern BI Accenture. This allows external modeling of roles via group membership. You know your needs access to a period of values should start in a schema hbase is there is apache license keys. Because tunings and construct and use cases, or restoring a procedural language for virtual database checking methods to region server is very fast and attributes. BigTable Rutgers CS. Let us look at the data model of HBASE and then understand the implementation architecture. Spark Schema defines the structure of the data if other words it simple the. In computing a school database GDB is a judge that uses graph structures for semantic queries with nodes edges and properties to represent and store data between key output of every system also the graph or interior or relationship The graph relates the data items in and store until a collection of nodes and. -

Dissertation Enabling Model-Driven Live Analytics for Cyber-Physical
PhD-FSTC-2016-43 The Faculty of Science, Technology and Communication Dissertation Defence held on 08/11/2016 in Luxembourg to obtain the degree of Docteur de l’Université du Luxembourg en Informatique by Thomas Hartmann Born on 13th November 1981 in Kaufbeuren (Germany) Enabling Model-Driven Live Analytics For Cyber-Physical Systems: The Case of Smart Grids Dissertation defence committee Prof. Dr. Nicolas Navet, chairman Professor, University of Luxembourg, Luxembourg, Luxembourg Dr. François Fouquet, vice-chairman Research Associate, University of Luxembourg, Luxembourg, Luxembourg Prof. Dr. Yves Le Traon, supervisor Professor, University of Luxembourg, Luxembourg, Luxembourg Prof. Dr. Jordi Cabot, member Professor, Universitat Oberta de Catalunya, Castelldefels (Barcelona), Spain Prof. Dr. François Taïani, member Professor, Université de Rennes 1, Rennes Cedex, France Dr. Jacques Klein, expert Senior Research Scientist, University of Luxembourg, Luxembourg, Luxembourg Abstract Advances in software, embedded computing, sensors, and networking technologies will lead to a new generation of smart cyber-physical systems that will far exceed the capa- bilities of today's embedded systems. They will be entrusted with increasingly complex tasks like controlling electric grids or autonomously driving cars. These systems have the potential to lay the foundations for tomorrow's critical infrastructures, to form the basis of emerging and future smart services, and to improve the quality of our everyday lives in many areas. In order to solve their tasks, they have to continuously monitor and collect data from physical processes, analyse this data, and make decisions based on it. Making smart decisions requires a deep understanding of the environment, in- ternal state, and the impacts of actions. -

Amazon EMR Migration Guide How to Move Apache Spark and Apache Hadoop from On-Premises to AWS
Amazon EMR Migration Guide How to Move Apache Spark and Apache Hadoop From On-Premises to AWS December 2, 2020 Notices Customers are responsible for making their own independent assessment of the information in this document. This document: (a) is for informational purposes only, (b) represents current AWS product offerings and practices, which are subject to change without notice, and (c) does not create any commitments or assurances from AWS and its affiliates, suppliers or licensors. AWS products or services are provided “as is” without warranties, representations, or conditions of any kind, whether express or implied. The responsibilities and liabilities of AWS to its customers are controlled by AWS agreements, and this document is not part of, nor does it modify, any agreement between AWS and its customers. © 2020 Amazon Web Services, Inc. or its affiliates. All rights reserved. Contents Overview .............................................................................................................................. 1 Starting Your Journey .......................................................................................................... 3 Migration Approaches ...................................................................................................... 3 Prototyping ....................................................................................................................... 6 Choosing a Team ............................................................................................................