Donkey Orchid Symptomless Virus: a Viral 'Platypus'
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Native Orchid Society of South Australia
NATIVE ORCHID SOCIETY of SOUTH AUSTRALIA NATIVE ORCHID SOCIETY OF SOUTH AUSTRALIA JOURNAL Volume 6, No. 10, November, 1982 Registered by Australia Post Publication No. SBH 1344. Price 40c PATRON: Mr T.R.N. Lothian PRESIDENT: Mr J.T. Simmons SECRETARY: Mr E.R. Hargreaves 4 Gothic Avenue 1 Halmon Avenue STONYFELL S.A. 5066 EVERARD PARK SA 5035 Telephone 32 5070 Telephone 293 2471 297 3724 VICE-PRESIDENT: Mr G.J. Nieuwenhoven COMMITTFE: Mr R. Shooter Mr P. Barnes TREASURER: Mr R.T. Robjohns Mrs A. Howe Mr R. Markwick EDITOR: Mr G.J. Nieuwenhoven NEXT MEETING WHEN: Tuesday, 23rd November, 1982 at 8.00 p.m. WHERE St. Matthews Hail, Bridge Street, Kensington. SUBJECT: This is our final meeting for 1982 and will take the form of a Social Evening. We will be showing a few slides to start the evening. Each member is requested to bring a plate. Tea, coffee, etc. will be provided. Plant Display and Commentary as usual, and Christmas raffle. NEW MEMBERS Mr. L. Field Mr. R.N. Pederson Mr. D. Unsworth Mrs. P.A. Biddiss Would all members please return any outstanding library books at the next meeting. FIELD TRIP -- CHANGE OF DATE AND VENUE The Field Trip to Peters Creek scheduled for 27th November, 1982, and announced in the last Journal has been cancelled. The extended dry season has not been conducive to flowering of the rarer moisture- loving Microtis spp., which were to be the objective of the trip. 92 FIELD TRIP - CHANGE OF DATE AND VENUE (Continued) Instead, an alternative trip has been arranged for Saturday afternoon, 4th December, 1982, meeting in Mount Compass at 2.00 p.m. -

Actes Du 15E Colloque Sur Les Orchidées De La Société Française D’Orchidophilie
Cah. Soc. Fr. Orch., n° 7 (2010) – Actes 15e colloque de la Société Française d’Orchidophilie, Montpellier Actes du 15e colloque sur les Orchidées de la Société Française d’Orchidophilie du 30 mai au 1er juin 2009 Montpellier, Le Corum Comité d’organisation : Daniel Prat, Francis Dabonneville, Philippe Feldmann, Michel Nicole, Aline Raynal-Roques, Marc-Andre Selosse, Bertrand Schatz Coordinateurs des Actes Daniel Prat & Bertrand Schatz Affiche du Colloque : Conception : Francis Dabonneville Photographies de Francis Dabonneville & Bertrand Schatz Cahiers de la Société Française d’Orchidophilie, N° 7, Actes du 15e Colloque sur les orchidées de la Société Française d’Orchidophilie. ISSN 0750-0386 © SFO, Paris, 2010 Certificat d’inscription à la commission paritaire N° 55828 ISBN 978-2-905734-17-4 Actes du 15e colloque sur les Orchidées de la Société Française d’Orchidophilie, D. Prat et B. Schatz, Coordinateurs, SFO, Paris, 2010, 236 p. Société Française d’Orchidophilie 17 Quai de la Seine, 75019 Paris Cah. Soc. Fr. Orch., n° 7 (2010) – Actes 15e colloque de la Société Française d’Orchidophilie, Montpellier Préface Ce 15e colloque marque le 40e anniversaire de notre société, celle-ci ayant vu le jour en 1969. Notre dernier colloque se tenait il y a 10 ans à Paris en 1999, 10 ans c’est long, 10 ans c’est très loin. Il fallait que la SFO renoue avec cette traditionnelle organisation de colloques, manifestation qui a contribué à lui accorder la place prépondérante qu’elle occupe au sein des orchidophiles français et de la communauté scientifique. C’est chose faite aujourd’hui. Nombreux sont les thèmes qui font l’objet de communications par des intervenants dont les compétences dans le domaine de l’orchidologie ne sont plus à prouver. -

Characterization of P1 Leader Proteases of the Potyviridae Family
Characterization of P1 leader proteases of the Potyviridae family and identification of the host factors involved in their proteolytic activity during viral infection Hongying Shan Ph.D. Dissertation Madrid 2018 UNIVERSIDAD AUTONOMA DE MADRID Facultad de Ciencias Departamento de Biología Molecular Characterization of P1 leader proteases of the Potyviridae family and identification of the host factors involved in their proteolytic activity during viral infection Hongying Shan This thesis is performed in Departamento de Genética Molecular de Plantas of Centro Nacional de Biotecnología (CNB-CSIC) under the supervision of Dr. Juan Antonio García and Dr. Bernardo Rodamilans Ramos Madrid 2018 Acknowledgements First of all, I want to express my appreciation to thesis supervisors Bernardo Rodamilans and Juan Antonio García, who gave the dedicated guidance to this thesis. I also want to say thanks to Carmen Simón-Mateo, Fabio Pasin, Raquel Piqueras, Beatriz García, Mingmin, Zhengnan, Wenli, Linlin, Ruiqiang, Runhong and Yuwei, who helped me and provided interesting suggestions for the thesis as well as technical support. Thanks to the people in the greenhouse (Tomás Heras, Alejandro Barrasa and Esperanza Parrilla), in vitro plant culture facility (María Luisa Peinado and Beatriz Casal), advanced light microscopy (Sylvia Gutiérrez and Ana Oña), photography service (Inés Poveda) and proteomics facility (Sergio Ciordia and María Carmen Mena). Thanks a lot to all the assistance from lab313 colleagues. Thanks a lot to the whole CNB. Thanks a lot to the Chinese Scholarship Council. Thanks a lot to all my friends. Thanks a lot to my family. Madrid 20/03/2018 Index I CONTENTS Abbreviations………………………………………….……………………….……...VII Viruses cited…………………………………………………………………..……...XIII Summary…………………………………………………………………...….…….XVII Resumen…………………………………………………………......…...…………..XXI I. -

Nucleotide Sequence and Phylogenetic Analysis of a Bamboo Mosaic Potexvirus Isolate from Common Bamboo (Bambusa Vulgaris Mcclure)
YangBot. Bull. et al. Acad. Nucleotide Sin. (1997) sequence 38: 77-84 of BaMV-V RNA 77 Nucleotide sequence and phylogenetic analysis of a bamboo mosaic potexvirus isolate from common bamboo (Bambusa vulgaris McClure) Chi-Chen Yang1,2, Jih-Shiou Liu1, Chan-Pin Lin2 and Na-Sheng Lin1,3 1Institute of Botany, Academia Sinica, Taipei, Taiwan 115, Republic of China 2Department of Plant Pathology and Entomology, National Taiwan University, Taipei, Taiwan, Republic of China (Received September 20, 1996; Accepted January 30, 1997) Abstract. The complete cDNA sequence corresponding to the genomic RNA of an isolate bamboo mosaic potexvirus (BaMV-V) from common bamboo (Bambusa vulgaris McClure) was determined. This isolate is the first potexvirus with which a satellite RNA has been associated. The genome organization of BaMV-V, similar to those of other potexviruses, contained five open reading frames (ORFs 15) coding for polypeptides with molecular weight of 156 kDa, 28 kDa, 13 kDa, 6 kDa, and 25 kDa, respectively. Nucleotide sequence analysis showed a 10.0% difference from the BaMV-O isolate previously described whereas the amino acid comparison showed a difference of 3.2%. When three conservative domains of RNA dependent RNA polymerase (RdRp) were used for phylogenetic analysis, the greatest variation between two strains of each virus was only 12.8% of that between the two closest members of the potexvirus group. The grouping of potexviruses distinct from other groups of plant viruses was also confirmed by a comparision of three conservative motifs of RdRp. Keywords: Bamboo mosaic virus; Nucleotide sequence; Potexvirus. Introduction virus M, PVM) (Zavriev et al., 1991), hordeivirus (e.g. -

Tamada-Text R3 HK-TT
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Okayama University Scientific Achievement Repository Tamada & Kondo - 1 Journal of General Plant Pathology Biological and genetic diversity of plasmodiophorid-transmitted viruses and their vectors Tetsuo Tamada* and Hideki Kondo Institute of Plant Science and Resources (IPSR), Okayama University, Kurashiki 710-0046, Japan. Current address for T. Tamada: Kita 10, Nishi 1, 13-2-606, Sapporo, 001-0010, Japan. *Corresponding author: Tetsuo Tamada; E-mail: [email protected] Total text pages 32 Word counts: 10 words (title); 147 words (Abstract); 6004 words (Main Text) Tables: 3; Figure: 1; Supplementary figure: 1 Tamada & Kondo - 2 Abstract About 20 species of viruses belonging to five genera, Benyvirus, Furovirus, Pecluvirus, Pomovirus and Bymovirus, are known to be transmitted by plasmodiophorids. These viruses have all positive-sense single-stranded RNA genomes that consist of two to five RNA components. Three species of plasmodiophorids are recognized as vectors: Polymyxa graminis, P. betae, and Spongospora subterranea. The viruses can survive in soil within the long-lived resting spores of the vector. There are biological and genetic variations in both virus and vector species. Many of the viruses have become the causal agents of important diseases in major crops, such as rice, wheat, barley, rye, sugar beet, potato, and groundnut. Control measure is dependent on the development of the resistant cultivars. During the last half a century, several virus diseases have been rapidly spread and distributed worldwide. For the six major virus diseases, their geographical distribution, diversity, and genetic resistance are addressed. -

The Development and Characterisation of Grapevine Virus-Based
The development and characterisation of grapevine virus-based expression vectors by Jacques du Preez Presented in partial fulfilment of the requirements for the degree Doctor of Philosophy at the Department of Genetics, Stellenbosch University March 2010 Supervisors: Prof JT Burger and Dr DE Goszczynski Study leader: Dr D Stephan Declaration I, the undersigned, hereby declare that the work contained in this thesis is my own original work and that I have not previously in its entirety or in part submitted it at any university for a degree. ______________________ Date: _______________ Jacques du Preez Copyright © 2010 Stellenbosch University All rights reserved ii Abstract Grapevine ( Vitis vinifera L.) is a very important agricultural commodity that needs to be protected. To achieve this several in vivo tools are needed for the study of this crop and the pathogens that infect it. Recently the grapevine genome has been sequenced and the next important step will be gene annotation and function using these in vivo tools. In this study the use of Grapevine virus A (GVA), genus Vitivirus , family Flexiviridae , as transient expression and VIGS vector for heterologous protein expression and functional genomics in Nicotiana benthamiana and V . vinifera were evaluated. Full-length genomic sequences of three South African variants of the virus (GTR11, GTG111 and GTR12) were generated and used in a molecular sequence comparison study. Results confirmed the separation of GVA variants into three groups, with group III (mild variants) being the most distantly related. It showed the high molecular heterogeneity of the virus and that ORF 2 was the most diverse. The GVA variants GTG111, GTR12 and GTR11 were placed in molecular groups I, II and III respectively. -

Disruption of Virus Movement Confers Broad-Spectrum Resistance Against
Proc. Nati. Acad. Sci. USA Vol. 91, pp. 10310-10314, October 1994 Plant Biology Disruption of virus movement confers broad-spectrum resistance against systemic infection by plant viruses with a triple gene block (trnenic plant/doinat negative mutaton/ n l movement proein) DAVID L. BECK, CRAIG J. VAN DOLLEWEERD, TONY J. LOUGH, EZEQUIEL BALMORI, DAVIN M. VOOT, MARK T. ANDERSEN, IONA E. W. O'BRIEN, AND RICHARD L. S. FORSTERt Molecular Genetics Group, The Horticultural and Food Research Institute of New Zealand Ltd., Private Bag 92169, Auckland, New Zealand Communicated by George Bruening, June 23, 1994 ABSTRACT White clover mosaic virus strain 0 (WCIMV- tially difficult. New forms ofresistance active against several 0), species of the Potexvirus genus, contains a set of three different viruses or groups of viruses are being sought. One partially overlapping genes (the triple gene block) that encodes such approach involved the introduction of the gene coding nonvirion proteins of 26 kDa, 13 kDa, and 7 kDa. These for rat 2'-5' oligoadenylate synthetase into the genome of proteins are necesy for cell-to-cell movement in plants but potato plants (4). Genetically engineering transgenic plants to not for replication. The WCIMV-O 13-kDa gene was mutated block virus movement, mimicking the mechanism of some (to 13*) in a region of the gene that is conserved in all viruses natural resistance genes, has been proposed (1, 5, 6) but known to possess triple-gene-block proteins. All 10 13* trans- remains mostly unexploited. genic lines of Nicodiana benthamiana designed to express the The movement function of plant viruses can be comple- mutated movement protein were shown to be resistant to mented by another, frequently unrelated, virus in double systemic infection by WCIMV-O at 1 jug ofWCIMV virions per infections (7). -
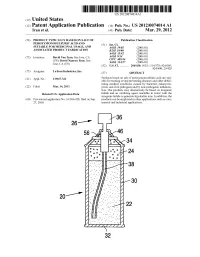
Us 2012/0074.014 A1 2 .. 1
US 2012O074O14A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2012/0074.014 A1 Tran et al. (43) Pub. Date: Mar. 29, 2012 (54) PRODUCT TYPICALLY BASED ON SALT OF Publication Classification PEROXYMONOSULFURIC ACID AND (51) Int. Cl SUITABLE FOR MEDICINAL USAGE, AND iBio/02 (2006.01) ASSOCATED PRODUCT FABRICATION B23P 19/00 (2006.01) A633/42 (2006.01) (75) Inventors: David Van Tran, San Jose, CA A6IR 9/14 (2006.01) (US); David Nguyen Tran, San CD7C 409/244 (2006.01) Jose, CA (US) A6II 3/327 (2006.01) s (52) U.S. Cl. ............. 206/438: 562/1; 514/578; 424/605; 424/400; 29/428 (73) Assignee: LuTran Industries, Inc. (57) ABSTRACT 21) Appl. No.: 13AO47,742 Products based on salt of peroxyperoxVmonosulfuric acid are suit (21) Appl. No 9 able for treating or/and preventing diseases and other debili tating medical conditions caused by bacterial, eukaryotic, (22) Filed: Mar. 14, 2011 prion, and viral pathogens and by non-pathogenic inflamma tion. The products may alternatively be based on inorganic Related U.S. Application Data halide and an oxidizing agent reactable in water with the inorganic halide to generate hypohalite ions. In addition, the (60) Provisional application No. 61/386,928, filed on Sep. products can be employed in other applications such as com 27, 2010. mercial and industrial applications. 5T. 2 .. 2. 1.4. J., Patent Application Publication Mar. 29, 2012 Sheet 1 of 2 US 2012/0074014 A1 cro Q-D 22 Fig. 2a Fig.2b Patent Application Publication Mar. 29, 2012 Sheet 2 of 2 US 2012/0074014 A1 90 92 US 2012/0074014 A1 Mar. -

Evidence to Support Safe Return to Clinical Practice by Oral Health Professionals in Canada During the COVID-19 Pandemic: a Repo
Evidence to support safe return to clinical practice by oral health professionals in Canada during the COVID-19 pandemic: A report prepared for the Office of the Chief Dental Officer of Canada. November 2020 update This evidence synthesis was prepared for the Office of the Chief Dental Officer, based on a comprehensive review under contract by the following: Paul Allison, Faculty of Dentistry, McGill University Raphael Freitas de Souza, Faculty of Dentistry, McGill University Lilian Aboud, Faculty of Dentistry, McGill University Martin Morris, Library, McGill University November 30th, 2020 1 Contents Page Introduction 3 Project goal and specific objectives 3 Methods used to identify and include relevant literature 4 Report structure 5 Summary of update report 5 Report results a) Which patients are at greater risk of the consequences of COVID-19 and so 7 consideration should be given to delaying elective in-person oral health care? b) What are the signs and symptoms of COVID-19 that oral health professionals 9 should screen for prior to providing in-person health care? c) What evidence exists to support patient scheduling, waiting and other non- treatment management measures for in-person oral health care? 10 d) What evidence exists to support the use of various forms of personal protective equipment (PPE) while providing in-person oral health care? 13 e) What evidence exists to support the decontamination and re-use of PPE? 15 f) What evidence exists concerning the provision of aerosol-generating 16 procedures (AGP) as part of in-person -

080058-92.02.002.Pdf
'Jeqruq ',{ueq1y 'pusD{u"rd ,Isad sed,{1i(1runuruoc Jeqlo ss sB JoJ pqse^]sq pua {rsrruaq II I I[€^\ 'eprsaeo eq o1 ,(1e41 s4serc! (roloctsp^tp mdQocng) IuJa4 4ru"Jg ol JeuB€raql lsEe-qlnos-ls"e sJapuDerl 11e 'eepJ?I ',{ellBeq^{ sapnlcu! 1 esnecaq uorleSgsa,rur aaJs puB ol qlnos ol lsue'dnuuBN ot Jo Fl.qdBJSoeB erll sB ueJJ8^\ eql esoqc e/l\ lsorrrl€ r{uou anp spuoq l! eJaq,r Je^ry ,([auuoq aqt ol lculstpqns lBcruslog ut"ld Je^rU gocs egl do1 eq1 sso:cr '1sare1m Jo NRo-r{lnos-ls"e luraueSelour Jo eq oslr ueql 'ralry poo.r$lt?tg eq1 uo e8pug repusxelv ol ,{ru 1sq 1ua1dprru qJJBeseJcrlsuou Jo ,l\erleJ aqJ lsua-qlnos-g$os sun-r ifurprmoq prrvJu Jo rueqUou aql 'sn[L '(SS 'd 'uousrrrJoJr[ dn8uJlpl urorJ :OgOt preag) lueuoduoc cglualJs elqs[r8^? lseq luucgruErs € eq ol sesrea JoloJtslaqp e:eql\ eq1 q1m,(4snpw drqcpoo,{ aq1 eEeuuu pnB rolmour ? 'lueue8su"l u.{erp sr,{:epunoq ruaquou eql elq,t\ 'lse/rt pu qlnos IrlSru 11 leqf os I purl pu? uorts^lesuoC 'lseoc sgl ol "es aq1 .{q paprmoq sr lI qmos eqt uo Jo lueulredaq eqt ,{q pelJnpuoJ Bllerlsny {*qlv ot e8pJg alsqumlulq-urrnoe-I eqt uo dn?u1ge1 lsea-rllnos u1 luerua8yuvu lseJoJ Jo lcedur eq1 uo qcJBoser uo.5 u{ 0o€ re^o spuelxe lcFlslpgns uerr"d\ eql Jo ?rterler B ol uorpquluoa v sa ,{luzuud esoP s3^\ IroA\ eqJ qcJReseJamlry ro3 sequoud 'errqsadsred 'EcefoJd luuor8a.r e ur seulluepr puE rIcJBeseJ tuaJrnc ol sJeJeJ ur"{ ol pernJuoo queuodwoc esoql go 'BroU '(9961 lseloJ lueuecqd r"lncsBl u,llou{ eql 3o 1s1 e quaserd 11 s8q lculslPgns uerrB eloq^1 eql JoJ P€lqBua \ p-rceg rnras) lclrtspgns lBcruslog -

Molecular Analysis of Vanilla Mosaic Virus from the Cook Islands
Molecular Analysis of Vanilla mosaic virus from the Cook Islands Christopher Puli’uvea A thesis submitted to Auckland University of Technology in partial fulfilment of the requirements for the degree of Master of Science (MSc) 2017 School of Science I Abstract Vanilla was first introduced to French Polynesia in 1848 and from 1899-1966 was a major export for French Polynesia who then produced an average of 158 tonnes of cured Vanilla tahitensis beans annually. In 1967, vanilla production declined rapidly to a low of 0.6 tonnes by 1981, which prompted a nation-wide investigation with the aim of restoring vanilla production to its former levels. As a result, a mosaic-inducing virus was discovered infecting V. tahitensis that was distinct from Cymbidium mosaic virus (CyMV) and Odontoglossum ringspot virus (ORSV) but serologically related to dasheen mosaic virus (DsMV). The potyvirus was subsequently named vanilla mosaic virus (VanMV) and was later reported to infect V. tahitensis in the Cook Islands and V. planifolia in Fiji and Vanuatu. Attempts were made to mechanically inoculate VanMV to a number of plants that are susceptible to DsMV, but with no success. Based on a partial sequence analysis, VanMV-FP (French Polynesian isolate) and VanMV-CI (Cook Islands isolate) were later characterised as strains of DsMV exclusively infecting vanilla. Since its discovery, little information is known about how VanMV-CI acquired the ability to exclusively infect vanilla and lose its ability to infect natural hosts of DsMV or vice versa. The aims of this research were to characterise the VanMV genome and attempt to determine the molecular basis for host range specificity of VanMV-CI. -

Viroze Biljaka 2010
VIROZE BILJAKA Ferenc Bagi Stevan Jasnić Dragana Budakov Univerzitet u Novom Sadu, Poljoprivredni fakultet Novi Sad, 2016 EDICIJA OSNOVNI UDŽBENIK Osnivač i izdavač edicije Univerzitet u Novom Sadu, Poljoprivredni fakultet Trg Dositeja Obradovića 8, 21000 Novi Sad Godina osnivanja 1954. Glavni i odgovorni urednik edicije Dr Nedeljko Tica, redovni profesor Dekan Poljoprivrednog fakulteta Članovi komisije za izdavačku delatnost Dr Ljiljana Nešić, vanredni profesor – predsednik Dr Branislav Vlahović, redovni profesor – član Dr Milica Rajić, redovni profesor – član Dr Nada Plavša, vanredni profesor – član Autori dr Ferenc Bagi, vanredni profesor dr Stevan Jasnić, redovni profesor dr Dragana Budakov, docent Glavni i odgovorni urednik Dr Nedeljko Tica, redovni profesor Dekan Poljoprivrednog fakulteta u Novom Sadu Urednik Dr Vera Stojšin, redovni profesor Direktor departmana za fitomedicinu i zaštitu životne sredine Recenzenti Dr Vera Stojšin, redovni profesor, Univerzitet u Novom Sadu, Poljoprivredni fakultet Dr Mira Starović, naučni savetnik, Institut za zaštitu bilja i životnu sredinu, Beograd Grafički dizajn korice Lea Bagi Izdavač Univerzitet u Novom Sadu, Poljoprivredni fakultet, Novi Sad Zabranjeno preštampavanje i fotokopiranje. Sva prava zadržava izdavač. ISBN 978-86-7520-372-8 Štampanje ovog udžbenika odobrilo je Nastavno-naučno veće Poljoprivrednog fakulteta u Novom Sadu na sednici od 11. 07. 2016.godine. Broj odluke 1000/0102-797/9/1 Tiraž: 20 Mesto i godina štampanja: Novi Sad, 2016. CIP - Ʉɚɬɚɥɨɝɢɡɚɰɢʁɚɭɩɭɛɥɢɤɚɰɢʁɢ ȻɢɛɥɢɨɬɟɤɚɆɚɬɢɰɟɫɪɩɫɤɟɇɨɜɢɋɚɞ