Tamada-Text R3 HK-TT
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Molecular Identification of Fungi
Molecular Identification of Fungi Youssuf Gherbawy l Kerstin Voigt Editors Molecular Identification of Fungi Editors Prof. Dr. Youssuf Gherbawy Dr. Kerstin Voigt South Valley University University of Jena Faculty of Science School of Biology and Pharmacy Department of Botany Institute of Microbiology 83523 Qena, Egypt Neugasse 25 [email protected] 07743 Jena, Germany [email protected] ISBN 978-3-642-05041-1 e-ISBN 978-3-642-05042-8 DOI 10.1007/978-3-642-05042-8 Springer Heidelberg Dordrecht London New York Library of Congress Control Number: 2009938949 # Springer-Verlag Berlin Heidelberg 2010 This work is subject to copyright. All rights are reserved, whether the whole or part of the material is concerned, specifically the rights of translation, reprinting, reuse of illustrations, recitation, broadcasting, reproduction on microfilm or in any other way, and storage in data banks. Duplication of this publication or parts thereof is permitted only under the provisions of the German Copyright Law of September 9, 1965, in its current version, and permission for use must always be obtained from Springer. Violations are liable to prosecution under the German Copyright Law. The use of general descriptive names, registered names, trademarks, etc. in this publication does not imply, even in the absence of a specific statement, that such names are exempt from the relevant protective laws and regulations and therefore free for general use. Cover design: WMXDesign GmbH, Heidelberg, Germany, kindly supported by ‘leopardy.com’ Printed on acid-free paper Springer is part of Springer Science+Business Media (www.springer.com) Dedicated to Prof. Lajos Ferenczy (1930–2004) microbiologist, mycologist and member of the Hungarian Academy of Sciences, one of the most outstanding Hungarian biologists of the twentieth century Preface Fungi comprise a vast variety of microorganisms and are numerically among the most abundant eukaryotes on Earth’s biosphere. -

Nucleotide Sequence and Phylogenetic Analysis of a Bamboo Mosaic Potexvirus Isolate from Common Bamboo (Bambusa Vulgaris Mcclure)
YangBot. Bull. et al. Acad. Nucleotide Sin. (1997) sequence 38: 77-84 of BaMV-V RNA 77 Nucleotide sequence and phylogenetic analysis of a bamboo mosaic potexvirus isolate from common bamboo (Bambusa vulgaris McClure) Chi-Chen Yang1,2, Jih-Shiou Liu1, Chan-Pin Lin2 and Na-Sheng Lin1,3 1Institute of Botany, Academia Sinica, Taipei, Taiwan 115, Republic of China 2Department of Plant Pathology and Entomology, National Taiwan University, Taipei, Taiwan, Republic of China (Received September 20, 1996; Accepted January 30, 1997) Abstract. The complete cDNA sequence corresponding to the genomic RNA of an isolate bamboo mosaic potexvirus (BaMV-V) from common bamboo (Bambusa vulgaris McClure) was determined. This isolate is the first potexvirus with which a satellite RNA has been associated. The genome organization of BaMV-V, similar to those of other potexviruses, contained five open reading frames (ORFs 15) coding for polypeptides with molecular weight of 156 kDa, 28 kDa, 13 kDa, 6 kDa, and 25 kDa, respectively. Nucleotide sequence analysis showed a 10.0% difference from the BaMV-O isolate previously described whereas the amino acid comparison showed a difference of 3.2%. When three conservative domains of RNA dependent RNA polymerase (RdRp) were used for phylogenetic analysis, the greatest variation between two strains of each virus was only 12.8% of that between the two closest members of the potexvirus group. The grouping of potexviruses distinct from other groups of plant viruses was also confirmed by a comparision of three conservative motifs of RdRp. Keywords: Bamboo mosaic virus; Nucleotide sequence; Potexvirus. Introduction virus M, PVM) (Zavriev et al., 1991), hordeivirus (e.g. -

Phylogenomics Supports the Monophyly of the Cercozoa T ⁎ Nicholas A.T
Molecular Phylogenetics and Evolution 130 (2019) 416–423 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev Phylogenomics supports the monophyly of the Cercozoa T ⁎ Nicholas A.T. Irwina, , Denis V. Tikhonenkova,b, Elisabeth Hehenbergera,1, Alexander P. Mylnikovb, Fabien Burkia,2, Patrick J. Keelinga a Department of Botany, University of British Columbia, Vancouver V6T 1Z4, British Columbia, Canada b Institute for Biology of Inland Waters, Russian Academy of Sciences, Borok 152742, Russia ARTICLE INFO ABSTRACT Keywords: The phylum Cercozoa consists of a diverse assemblage of amoeboid and flagellated protists that forms a major Cercozoa component of the supergroup, Rhizaria. However, despite its size and ubiquity, the phylogeny of the Cercozoa Rhizaria remains unclear as morphological variability between cercozoan species and ambiguity in molecular analyses, Phylogeny including phylogenomic approaches, have produced ambiguous results and raised doubts about the monophyly Phylogenomics of the group. Here we sought to resolve these ambiguities using a 161-gene phylogenetic dataset with data from Single-cell transcriptomics newly available genomes and deeply sequenced transcriptomes, including three new transcriptomes from Aurigamonas solis, Abollifer prolabens, and a novel species, Lapot gusevi n. gen. n. sp. Our phylogenomic analysis strongly supported a monophyletic Cercozoa, and approximately-unbiased tests rejected the paraphyletic topologies observed in previous studies. The transcriptome of L. gusevi represents the first transcriptomic data from the large and recently characterized Aquavolonidae-Treumulida-'Novel Clade 12′ group, and phyloge- nomics supported its position as sister to the cercozoan subphylum, Endomyxa. These results provide insights into the phylogeny of the Cercozoa and the Rhizaria as a whole. -

Viruses Virus Diseases Poaceae(Gramineae)
Viruses and virus diseases of Poaceae (Gramineae) Viruses The Poaceae are one of the most important plant families in terms of the number of species, worldwide distribution, ecosystems and as ingredients of human and animal food. It is not surprising that they support many parasites including and more than 100 severely pathogenic virus species, of which new ones are being virus diseases regularly described. This book results from the contributions of 150 well-known specialists and presents of for the first time an in-depth look at all the viruses (including the retrotransposons) Poaceae(Gramineae) infesting one plant family. Ta xonomic and agronomic descriptions of the Poaceae are presented, followed by data on molecular and biological characteristics of the viruses and descriptions up to species level. Virus diseases of field grasses (barley, maize, rice, rye, sorghum, sugarcane, triticale and wheats), forage, ornamental, aromatic, wild and lawn Gramineae are largely described and illustrated (32 colour plates). A detailed index Sciences de la vie e) of viruses and taxonomic lists will help readers in their search for information. Foreworded by Marc Van Regenmortel, this book is essential for anyone with an interest in plant pathology especially plant virology, entomology, breeding minea and forecasting. Agronomists will also find this book invaluable. ra The book was coordinated by Hervé Lapierre, previously a researcher at the Institut H. Lapierre, P.-A. Signoret, editors National de la Recherche Agronomique (Versailles-France) and Pierre A. Signoret emeritus eae (G professor and formerly head of the plant pathology department at Ecole Nationale Supérieure ac Agronomique (Montpellier-France). Both have worked from the late 1960’s on virus diseases Po of Poaceae . -

Distribution of Two Formae Speciales of Polymyxa Graminis in the Czech Republic*
PLANT SCIENCES DISTRIBUTION OF TWO FORMAE SPECIALES OF POLYMYXA GRAMINIS IN THE CZECH REPUBLIC* L. Grimová, L. Winkowska, B. Špuláková, P. Růžičková, P. Ryšánek Czech University of Life Sciences Prague, Faculty of Agrobiology, Food and Natural Resources, Department of Crop Protection, Prague, Czech Republic It has been shown that two formae speciales of P. graminis, namely f. sp. temperata (ribotype Pg-I) and f. sp. tepida (ribotype Pg-II), are widely distributed throughout temperate areas of Europe. In this study, the presence of both forms of the temper- ate Polymyxa spp. was identified in soil samples from different locations of the Czech Republic during a survey performed in 2012 and 2013. Based on polymerase chain reaction results, of the total 58 tested samples, 67.2% contained at least one monitored forma specialis. Specifically, P. graminis f. sp. temperata was detected in 48.3% of soil samples, while P. graminis f. sp. tepida was detected in 44.8% of samples. Mixed populations were found in 25.9% of the tested areas. This plasmodio- phorid was confirmed not only in crop fields but also in meadows and forests in all explored regions. Our results extend the knowledge on the distribution of both ribotypes of P. graminis and provide the first evidence of f. sp. tepida within the Czech Republic. Plasmodiophoromycetes, temperate ribotypes, monitoring, PCR doi: 10.1515/sab-2017-0011 Received for publication on March 17, 2016 Accepted for publication on November 1, 2016 INTRODUCTION ous species in the Chenopodiaceae and the related plant families Amaranthaceae, Portulacaceae, and The genus Polymyxa represents one of ten genera in the Caryophyllaceae (Barr, Asher, 1992; Legrève family Plasmodiophoracea (order Plasmodiophorales, et al., 2002; R u s h , 2003). -

ICTV Code Assigned: 2011.001Ag Officers)
This form should be used for all taxonomic proposals. Please complete all those modules that are applicable (and then delete the unwanted sections). For guidance, see the notes written in blue and the separate document “Help with completing a taxonomic proposal” Please try to keep related proposals within a single document; you can copy the modules to create more than one genus within a new family, for example. MODULE 1: TITLE, AUTHORS, etc (to be completed by ICTV Code assigned: 2011.001aG officers) Short title: Change existing virus species names to non-Latinized binomials (e.g. 6 new species in the genus Zetavirus) Modules attached 1 2 3 4 5 (modules 1 and 9 are required) 6 7 8 9 Author(s) with e-mail address(es) of the proposer: Van Regenmortel Marc, [email protected] Burke Donald, [email protected] Calisher Charles, [email protected] Dietzgen Ralf, [email protected] Fauquet Claude, [email protected] Ghabrial Said, [email protected] Jahrling Peter, [email protected] Johnson Karl, [email protected] Holbrook Michael, [email protected] Horzinek Marian, [email protected] Keil Guenther, [email protected] Kuhn Jens, [email protected] Mahy Brian, [email protected] Martelli Giovanni, [email protected] Pringle Craig, [email protected] Rybicki Ed, [email protected] Skern Tim, [email protected] Tesh Robert, [email protected] Wahl-Jensen Victoria, [email protected] Walker Peter, [email protected] Weaver Scott, [email protected] List the ICTV study group(s) that have seen this proposal: A list of study groups and contacts is provided at http://www.ictvonline.org/subcommittees.asp . -

The Development and Characterisation of Grapevine Virus-Based
The development and characterisation of grapevine virus-based expression vectors by Jacques du Preez Presented in partial fulfilment of the requirements for the degree Doctor of Philosophy at the Department of Genetics, Stellenbosch University March 2010 Supervisors: Prof JT Burger and Dr DE Goszczynski Study leader: Dr D Stephan Declaration I, the undersigned, hereby declare that the work contained in this thesis is my own original work and that I have not previously in its entirety or in part submitted it at any university for a degree. ______________________ Date: _______________ Jacques du Preez Copyright © 2010 Stellenbosch University All rights reserved ii Abstract Grapevine ( Vitis vinifera L.) is a very important agricultural commodity that needs to be protected. To achieve this several in vivo tools are needed for the study of this crop and the pathogens that infect it. Recently the grapevine genome has been sequenced and the next important step will be gene annotation and function using these in vivo tools. In this study the use of Grapevine virus A (GVA), genus Vitivirus , family Flexiviridae , as transient expression and VIGS vector for heterologous protein expression and functional genomics in Nicotiana benthamiana and V . vinifera were evaluated. Full-length genomic sequences of three South African variants of the virus (GTR11, GTG111 and GTR12) were generated and used in a molecular sequence comparison study. Results confirmed the separation of GVA variants into three groups, with group III (mild variants) being the most distantly related. It showed the high molecular heterogeneity of the virus and that ORF 2 was the most diverse. The GVA variants GTG111, GTR12 and GTR11 were placed in molecular groups I, II and III respectively. -

Tamada-Text R3 HK-TT
Tamada & Kondo - 1 Journal of General Plant Pathology Biological and genetic diversity of plasmodiophorid-transmitted viruses and their vectors Tetsuo Tamada* and Hideki Kondo Institute of Plant Science and Resources (IPSR), Okayama University, Kurashiki 710-0046, Japan. Current address for T. Tamada: Kita 10, Nishi 1, 13-2-606, Sapporo, 001-0010, Japan. *Corresponding author: Tetsuo Tamada; E-mail: [email protected] Total text pages 32 Word counts: 10 words (title); 147 words (Abstract); 6004 words (Main Text) Tables: 3; Figure: 1; Supplementary figure: 1 Tamada & Kondo - 2 Abstract About 20 species of viruses belonging to five genera, Benyvirus, Furovirus, Pecluvirus, Pomovirus and Bymovirus, are known to be transmitted by plasmodiophorids. These viruses have all positive-sense single-stranded RNA genomes that consist of two to five RNA components. Three species of plasmodiophorids are recognized as vectors: Polymyxa graminis, P. betae, and Spongospora subterranea. The viruses can survive in soil within the long-lived resting spores of the vector. There are biological and genetic variations in both virus and vector species. Many of the viruses have become the causal agents of important diseases in major crops, such as rice, wheat, barley, rye, sugar beet, potato, and groundnut. Control measure is dependent on the development of the resistant cultivars. During the last half a century, several virus diseases have been rapidly spread and distributed worldwide. For the six major virus diseases, their geographical distribution, diversity, and genetic resistance are addressed. Keywords Soil-borne viruses; Benyvirus; Furovirus; Pecluvirus; Pomovirus; Bymovirus; Vector transmission; Plasmodiophorids; Polymyxa; Spongospora Introduction There is a group of viruses to be transmitted by olpidium and plasmodiophorid vectors, which is called soil-borne fungus-transmitted viruses. -

Small Hydrophobic Viral Proteins Involved in Intercellular Movement of Diverse Plant Virus Genomes Sergey Y
AIMS Microbiology, 6(3): 305–329. DOI: 10.3934/microbiol.2020019 Received: 23 July 2020 Accepted: 13 September 2020 Published: 21 September 2020 http://www.aimspress.com/journal/microbiology Review Small hydrophobic viral proteins involved in intercellular movement of diverse plant virus genomes Sergey Y. Morozov1,2,* and Andrey G. Solovyev1,2,3 1 A. N. Belozersky Institute of Physico-Chemical Biology, Moscow State University, Moscow, Russia 2 Department of Virology, Biological Faculty, Moscow State University, Moscow, Russia 3 Institute of Molecular Medicine, Sechenov First Moscow State Medical University, Moscow, Russia * Correspondence: E-mail: [email protected]; Tel: +74959393198. Abstract: Most plant viruses code for movement proteins (MPs) targeting plasmodesmata to enable cell-to-cell and systemic spread in infected plants. Small membrane-embedded MPs have been first identified in two viral transport gene modules, triple gene block (TGB) coding for an RNA-binding helicase TGB1 and two small hydrophobic proteins TGB2 and TGB3 and double gene block (DGB) encoding two small polypeptides representing an RNA-binding protein and a membrane protein. These findings indicated that movement gene modules composed of two or more cistrons may encode the nucleic acid-binding protein and at least one membrane-bound movement protein. The same rule was revealed for small DNA-containing plant viruses, namely, viruses belonging to genus Mastrevirus (family Geminiviridae) and the family Nanoviridae. In multi-component transport modules the nucleic acid-binding MP can be viral capsid protein(s), as in RNA-containing viruses of the families Closteroviridae and Potyviridae. However, membrane proteins are always found among MPs of these multicomponent viral transport systems. -

Disruption of Virus Movement Confers Broad-Spectrum Resistance Against
Proc. Nati. Acad. Sci. USA Vol. 91, pp. 10310-10314, October 1994 Plant Biology Disruption of virus movement confers broad-spectrum resistance against systemic infection by plant viruses with a triple gene block (trnenic plant/doinat negative mutaton/ n l movement proein) DAVID L. BECK, CRAIG J. VAN DOLLEWEERD, TONY J. LOUGH, EZEQUIEL BALMORI, DAVIN M. VOOT, MARK T. ANDERSEN, IONA E. W. O'BRIEN, AND RICHARD L. S. FORSTERt Molecular Genetics Group, The Horticultural and Food Research Institute of New Zealand Ltd., Private Bag 92169, Auckland, New Zealand Communicated by George Bruening, June 23, 1994 ABSTRACT White clover mosaic virus strain 0 (WCIMV- tially difficult. New forms ofresistance active against several 0), species of the Potexvirus genus, contains a set of three different viruses or groups of viruses are being sought. One partially overlapping genes (the triple gene block) that encodes such approach involved the introduction of the gene coding nonvirion proteins of 26 kDa, 13 kDa, and 7 kDa. These for rat 2'-5' oligoadenylate synthetase into the genome of proteins are necesy for cell-to-cell movement in plants but potato plants (4). Genetically engineering transgenic plants to not for replication. The WCIMV-O 13-kDa gene was mutated block virus movement, mimicking the mechanism of some (to 13*) in a region of the gene that is conserved in all viruses natural resistance genes, has been proposed (1, 5, 6) but known to possess triple-gene-block proteins. All 10 13* trans- remains mostly unexploited. genic lines of Nicodiana benthamiana designed to express the The movement function of plant viruses can be comple- mutated movement protein were shown to be resistant to mented by another, frequently unrelated, virus in double systemic infection by WCIMV-O at 1 jug ofWCIMV virions per infections (7). -
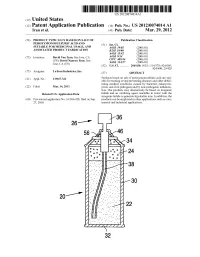
Us 2012/0074.014 A1 2 .. 1
US 2012O074O14A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2012/0074.014 A1 Tran et al. (43) Pub. Date: Mar. 29, 2012 (54) PRODUCT TYPICALLY BASED ON SALT OF Publication Classification PEROXYMONOSULFURIC ACID AND (51) Int. Cl SUITABLE FOR MEDICINAL USAGE, AND iBio/02 (2006.01) ASSOCATED PRODUCT FABRICATION B23P 19/00 (2006.01) A633/42 (2006.01) (75) Inventors: David Van Tran, San Jose, CA A6IR 9/14 (2006.01) (US); David Nguyen Tran, San CD7C 409/244 (2006.01) Jose, CA (US) A6II 3/327 (2006.01) s (52) U.S. Cl. ............. 206/438: 562/1; 514/578; 424/605; 424/400; 29/428 (73) Assignee: LuTran Industries, Inc. (57) ABSTRACT 21) Appl. No.: 13AO47,742 Products based on salt of peroxyperoxVmonosulfuric acid are suit (21) Appl. No 9 able for treating or/and preventing diseases and other debili tating medical conditions caused by bacterial, eukaryotic, (22) Filed: Mar. 14, 2011 prion, and viral pathogens and by non-pathogenic inflamma tion. The products may alternatively be based on inorganic Related U.S. Application Data halide and an oxidizing agent reactable in water with the inorganic halide to generate hypohalite ions. In addition, the (60) Provisional application No. 61/386,928, filed on Sep. products can be employed in other applications such as com 27, 2010. mercial and industrial applications. 5T. 2 .. 2. 1.4. J., Patent Application Publication Mar. 29, 2012 Sheet 1 of 2 US 2012/0074014 A1 cro Q-D 22 Fig. 2a Fig.2b Patent Application Publication Mar. 29, 2012 Sheet 2 of 2 US 2012/0074014 A1 90 92 US 2012/0074014 A1 Mar. -

Evidence to Support Safe Return to Clinical Practice by Oral Health Professionals in Canada During the COVID-19 Pandemic: a Repo
Evidence to support safe return to clinical practice by oral health professionals in Canada during the COVID-19 pandemic: A report prepared for the Office of the Chief Dental Officer of Canada. November 2020 update This evidence synthesis was prepared for the Office of the Chief Dental Officer, based on a comprehensive review under contract by the following: Paul Allison, Faculty of Dentistry, McGill University Raphael Freitas de Souza, Faculty of Dentistry, McGill University Lilian Aboud, Faculty of Dentistry, McGill University Martin Morris, Library, McGill University November 30th, 2020 1 Contents Page Introduction 3 Project goal and specific objectives 3 Methods used to identify and include relevant literature 4 Report structure 5 Summary of update report 5 Report results a) Which patients are at greater risk of the consequences of COVID-19 and so 7 consideration should be given to delaying elective in-person oral health care? b) What are the signs and symptoms of COVID-19 that oral health professionals 9 should screen for prior to providing in-person health care? c) What evidence exists to support patient scheduling, waiting and other non- treatment management measures for in-person oral health care? 10 d) What evidence exists to support the use of various forms of personal protective equipment (PPE) while providing in-person oral health care? 13 e) What evidence exists to support the decontamination and re-use of PPE? 15 f) What evidence exists concerning the provision of aerosol-generating 16 procedures (AGP) as part of in-person