The Tree of Life
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

North American Journal of Psychology, 1999. PUB DATE 1999-00-00 NOTE 346P.; Published Semi-Annually
DOCUMENT RESUME ED 449 388 CG 029 765 AUTHOR McCutcheon, Lynn E., Ed. TITLE North American Journal of Psychology, 1999. PUB DATE 1999-00-00 NOTE 346p.; Published semi-annually. AVAILABLE FROM NAJP, 240 Harbor Dr., Winter Garden, FL 34787 ($35 per annual subscription). Tel: 407-877-8364. PUB TYPE Collected Works Serials (022) JOURNAL CIT North American Journal of Psychology; vl n1-2 1999 EDRS PRICE MF01/PC14 Plus Postage. DESCRIPTORS *Psychology; *Research Tools; *Scholarly Journals; *Social Science Research ABSTRACT "North American Journal of Psychology" publishes scientific papers of general interest to psychologists and other social scientists. Articles included in volume 1 issue 1 (June 1999) are: "Generalist Looks at His Career in Teaching: Interview with Dr. Phil Zimbardo"; "Affective Information in Videos"; "Infant Communication"; "Defining Projective Techniques"; "Date Selection Choices in College Students"; "Study of the Personality of Violent Children"; "Behavioral and Institutional Theories of Human Resource Practices"; "Self-Estimates of Intelligence:"; "When the Going Gets Tough, the Tough Get Going"; "Behaviorism and Cognitivism in Learning Theory"; "On the Distinction between Behavioral Contagion, Conversion Conformity, and Compliance Conformity"; "Promoting Altruism in Troubled Youth"; The Influence of insecurity on Exchange and Communal Intimates"; "Height as Power in Women"; "Moderator Effects of Managerial Activity Inhibition on the Relation between Power versus Affiliation Motive Dominance and Econdmic Efficiency"; -

Foucault's Darwinian Genealogy
genealogy Article Foucault’s Darwinian Genealogy Marco Solinas Political Philosophy, University of Florence and Deutsches Institut Florenz, Via dei Pecori 1, 50123 Florence, Italy; [email protected] Academic Editor: Philip Kretsedemas Received: 10 March 2017; Accepted: 16 May 2017; Published: 23 May 2017 Abstract: This paper outlines Darwin’s theory of descent with modification in order to show that it is genealogical in a narrow sense, and that from this point of view, it can be understood as one of the basic models and sources—also indirectly via Nietzsche—of Foucault’s conception of genealogy. Therefore, this essay aims to overcome the impression of a strong opposition to Darwin that arises from Foucault’s critique of the “evolutionistic” research of “origin”—understood as Ursprung and not as Entstehung. By highlighting Darwin’s interpretation of the principles of extinction, divergence of character, and of the many complex contingencies and slight modifications in the becoming of species, this essay shows how his genealogical framework demonstrates an affinity, even if only partially, with Foucault’s genealogy. Keywords: Darwin; Foucault; genealogy; natural genealogies; teleology; evolution; extinction; origin; Entstehung; rudimentary organs “Our classifications will come to be, as far as they can be so made, genealogies; and will then truly give what may be called the plan of creation. The rules for classifying will no doubt become simpler when we have a definite object in view. We possess no pedigrees or armorial bearings; and we have to discover and trace the many diverging lines of descent in our natural genealogies, by characters of any kind which have long been inherited. -

Synthesis of Phylogeny and Taxonomy Into a Comprehensive Tree of Life
Synthesis of phylogeny and taxonomy into a comprehensive tree of life Cody E. Hinchliffa,1, Stephen A. Smitha,1,2, James F. Allmanb, J. Gordon Burleighc, Ruchi Chaudharyc, Lyndon M. Coghilld, Keith A. Crandalle, Jiabin Dengc, Bryan T. Drewf, Romina Gazisg, Karl Gudeh, David S. Hibbettg, Laura A. Katzi, H. Dail Laughinghouse IVi, Emily Jane McTavishj, Peter E. Midfordd, Christopher L. Owenc, Richard H. Reed, Jonathan A. Reesk, Douglas E. Soltisc,l, Tiffani Williamsm, and Karen A. Cranstonk,2 aEcology and Evolutionary Biology, University of Michigan, Ann Arbor, MI 48109; bInterrobang Corporation, Wake Forest, NC 27587; cDepartment of Biology, University of Florida, Gainesville, FL 32611; dField Museum of Natural History, Chicago, IL 60605; eComputational Biology Institute, George Washington University, Ashburn, VA 20147; fDepartment of Biology, University of Nebraska-Kearney, Kearney, NE 68849; gDepartment of Biology, Clark University, Worcester, MA 01610; hSchool of Journalism, Michigan State University, East Lansing, MI 48824; iBiological Science, Clark Science Center, Smith College, Northampton, MA 01063; jDepartment of Ecology and Evolutionary Biology, University of Kansas, Lawrence, KS 66045; kNational Evolutionary Synthesis Center, Duke University, Durham, NC 27705; lFlorida Museum of Natural History, University of Florida, Gainesville, FL 32611; and mComputer Science and Engineering, Texas A&M University, College Station, TX 77843 Edited by David M. Hillis, The University of Texas at Austin, Austin, TX, and approved July 28, 2015 (received for review December 3, 2014) Reconstructing the phylogenetic relationships that unite all line- published phylogenies are available only as journal figures, rather ages (the tree of life) is a grand challenge. The paucity of homologous than in electronic formats that can be integrated into databases and character data across disparately related lineages currently renders synthesis methods (7–9). -

Ten Misunderstandings About Evolution a Very Brief Guide for the Curious and the Confused by Dr
Ten Misunderstandings About Evolution A Very Brief Guide for the Curious and the Confused By Dr. Mike Webster, Dept. of Neurobiology and Behavior, Cornell Lab of Ornithology, Cornell University ([email protected]); February 2010 The current debate over evolution and “intelligent design” (ID) is being driven by a relatively small group of individuals who object to the theory of evolution for religious reasons. The debate is fueled, though, by misunderstandings on the part of the American public about what evolutionary biology is and what it says. These misunderstandings are exploited by proponents of ID, intentionally or not, and are often echoed in the media. In this booklet I briefly outline and explain 10 of the most common (and serious) misunderstandings. It is impossible to treat each point thoroughly in this limited space; I encourage you to read further on these topics and also by visiting the websites given on the resource sheet. In addition, I am happy to send a somewhat expanded version of this booklet to anybody who is interested – just send me an email to ask for one! What are the misunderstandings? 1. Evolution is progressive improvement of species Evolution, particularly human evolution, is often pictured in textbooks as a string of organisms marching in single file from “simple” organisms (usually a single celled organism or a monkey) on one side of the page and advancing to “complex” organisms on the opposite side of the page (almost invariably a human being). We have all seen this enduring image and likely have some version of it burned into our brains. -

The Consequences of Infelicity: the Effects of Unhappinesson
AN ABSTRACT OF THE THESIS OF Jorge R. Martinez for the degree of Master of Arts in Interdisciplinary Studies in Political Science, History and Foreign Languages and Literaturespresented on June 5, 1991. Title: The Consequences of Infelicity: The Effects of Unhappinesson Biological and Social Evolution Redacted for Privacy Abstract approved: Glen C. Dealy In social and biological evolution, infelicitycan operate as a driving motor to force change. In this essay, for life other than human, infelicity is equated with physical unfitness to compete for theresources of a specific niche. For humanity it is defined as the result ofan incongruity between a nation's culture and its government. The purpose of this study is to investigate how, for irrational life,unfitness can stimulate the creation of a new species and, for men, how the unhappiness of a nation may enhance its opportunity to enter a new socio-economic order. An evolutionary account about a possible way in which life could have evolved is offered, concentrating mainly on the transition from ape to a less remote ancestor of man, but also taking into consideration other life forms. Then, a parallel to social evolution is established. A study of the rise of capitalism in England, as well as the recent attempts to institute socialism in Latin America, are also explained as consequences of infelicity. c Copyright by Jorge R. Martinez June 5, 1991 All Rights Reserved The Consequences of Infelicity: the Effects of Unhappiness on Biological and Social Evolution by Jorge R. Martinez A THESIS -

Darwin's “Tree of Life”
Icons of Evolution? Why Much of What Jonathan Wells Writes about Evolution is Wrong Alan D. Gishlick, National Center for Science Education DARWIN’S “TREE OF LIFE” mon descent. Finally, he demands that text- books treat universal common ancestry as PHYLOGENETIC TREES unproven and refrain from illustrating that n biology, a phylogenetic tree, or phyloge- “theory” with misleading phylogenies. ny, is used to show the genealogic relation- Therefore, according to Wells, textbooks Iships of living things. A phylogeny is not should state that there is no evidence for com- so much evidence for evolution as much as it mon descent and that the most recent research is a codification of data about evolutionary his- refutes the concept entirely. Wells is complete- tory. According to biological evolution, organ- ly wrong on all counts, and his argument is isms share common ancestors; a phylogeny entirely based on misdirection and confusion. shows how organisms are related. The tree of He mixes up these various topics in order to life shows the path evolution took to get to the confuse the reader into thinking that when current diversity of life. It also shows that we combined, they show an endemic failure of can ascertain the genealogy of disparate living evolutionary theory. In effect, Wells plays the organisms. This is evidence for evolution only equivalent of an intellectual shell game, put- in that we can construct such trees at all. If ting so many topics into play that the “ball” of evolution had not happened or common ances- evolution gets lost. try were false, we would not be able to discov- THE CAMBRIAN EXPLOSION er hierarchical branching genealogies for ells claims that the Cambrian organisms (although textbooks do not general- Explosion “presents a serious chal- ly explain this well). -

The Rooting of the Universal Tree of Life Is Not Reliable
J Mol Evol (1999) 49:509–523 © Springer-Verlag New York Inc. 1999 The Rooting of the Universal Tree of Life Is Not Reliable Herve´ Philippe,1 Patrick Forterre2 1 Phyloge´nie et Evolution Mole´culaires (UPRESA 8080 CNRS), Baˆtiment 444, Universite´ Paris-Sud, 91405 Orsay-Cedex, France 2 Institut de Ge´ne´tique et Microbiologie (UMR 8621 CNRS), Baˆtiment 409, Universite´ Paris-Sud, 91405 Orsay-Cedex, France Abstract. Several composite universal trees connected terial rooting could be explained by an attraction be- by an ancestral gene duplication have been used to root tween this branch and the long branch of the outgroup. the universal tree of life. In all cases, this root turned out Finally, we suggested that an eukaryotic rooting could be to be in the eubacterial branch. However, the validity of a more fruitful working hypothesis, as it provides, for results obtained from comparative sequence analysis has example, a simple explanation to the high genetic simi- recently been questioned, in particular, in the case of larity of Archaebacteria and Eubacteria inferred from ancient phylogenies. For example, it has been shown that complete genome analysis. several eukaryotic groups are misplaced in ribosomal RNA or elongation factor trees because of unequal rates Key words: Root of the tree of life — ATPase — of evolution and mutational saturation. Furthermore, the Carbamoyl phosphate synthetase — Elongation factor — addition of new sequences to data sets has often turned tRNA synthetase — Signal recognition particle — Mu- apparently reasonable phylogenies into confused ones. tational saturation — Long branch attraction We have thus revisited all composite protein trees that have been used to root the universal tree of life up to now (elongation factors, ATPases, tRNA synthetases, car- bamoyl phosphate synthetases, signal recognition par- Introduction ticle proteins) with updated data sets. -

Reviews & Essays
Reviews & Essays scholars and much of the educated public Pinker the Prophet simply deny the good news. But prehistoric graves and records from twentieth-century By Robert Jervis hunter-gatherers reveal death rates due to warfare five to ten times that of modern Europe, and the homicide rate in Western Steven Pinker, The Better Angels of Our Na- Europe from 1300 to today has dropped ture: Why Violence Has Declined (New York: by a factor of between ten and fifty. When Viking Adult, 2011), 832 pp., $40.00. we read that after conquering a city the ancient Greeks killed all the men and sold ....ith the United States fighting the women and children into slavery, we ..two wars, countries from Tu- tend to let the phrases pass over us as we Wnisia to Syria either in or on the move on to admire Greek poetry, plays and brink of intrastate conflicts, bloodshed civilization. But this kind of slaughter was continuing in Sudan and reports that sui- central to the Greek way of life. cide bombers might foil airport security by Implicit throughout and explicit at the planting explosives within their bodies, it is very end is Pinker’s passionate belief that hard to be cheerful. But Harvard psycholo- contemporary attacks on the Enlighten- gist Steven Pinker tells us that we should ment and modernity are fundamentally be, that we are living in the least violent misguided. Critics often argue that material era ever. What’s more, he makes a case that and technical progress has been achieved will be hard to refute. -

Discover Biology, Develop Skills, and Make Connections
Discover Biology, Develop Skills, and Make Connections Since it’s trailblazing First Edition, Biological Sciences has delivered numerous biology teaching innovations that emphasize higher-order thinking skills and conceptual understanding rather than an encyclopedic grasp of what is known about biology. Central to this shift is a student-centered approach that provides support for mastering core content and developing skills that help students learn and practice biology. This model represents the . ultimately . to become . and then to apply overarching goal of the Seventh completing the course active learners what they have learned Edition: To help novice learners as expert learners who through practice . to new situations . progress from instruction . think like biologists. Instruction Practice Application Content Skills ThinkingT like a biologist A01_FREE8320_07_SE_FM.indd 1 11/21/18 12:01 AM Making Connections Through NEW Integrative End of Unit Case Study is introduced following Chapter 1. Each unit concludes with a 2-page spread that continues the story, guiding students through an exploration of key biological elements and scientific data. A unifying story about the evolutionary arms race between newts and garter snakes unfolds to illustrate how biology concepts and the various sub- disciplines of biology are connected across multiple levels from molecules, cells, and genetics to evolution and diversity, physiology, and ecology. Materials in Mastering Biology support in-class and out-of-class activities. END-OF-UNIT For media go to Mastering Biology ❚ CASE STUDY For an introduction to the Mystery of the Newt case study, see page 17. ••• Now that you’ve learned about evolutionary processes and patterns, it’s time to return Now that you have considered heritable variation Newt Snake UNIT 4 and fitness trade-os, the next step is to consider toxicity resistance to the Mystery of the Newt. -

Evidence for Evolution
CHAPTER 3 Evidence for Evolution VOLUTIONARY BIOLOGY HAS PROFOUNDLY altered our view of nature and of ourselves. At the beginning of this book, we showed the practical application of Eevolutionary biology to agriculture, biotechnology, and medicine. More broadly, evolutionary theory underpins all our knowledge of biology, explains how organisms came to be (both describing their history and identi- fying the processes that acted), and explains why they are as they are (why organisms reproduce sexually, why they age, and so on). How- ever, arguably its most important influence has been on how we view ourselves and our place in the world. The radical scope of evolution- ary biology has for many been hard to accept, and this has led to much misunderstanding and many objections. In this chapter, we summarize the evidence for evolution, clarify some common misun- derstandings, and discuss the wider implications of evolution by natural selection. Biological evolution was widely accepted soon after the publication of On the Origin of Species in 1859 (Chapter 1.x). Charles Darwin set out “one long argument” for the “descent with modification” of all liv- ing organisms, from one or a few common ancestors. He marshaled evidence from classification of organisms, from the fossil record, from geographic distribution of organisms, and by analogy with artificial se- lection. As we saw in Chapter 1, the detailed processes that cause evo- lution remained obscure until after the laws of heredity were established in the early 20th century. By the time of the Evolutionary Synthesis,in the mid-20th century, these processes were well understood and, cru- cially, it was established that adaptation is due to natural selection (Chapter 1.x). -
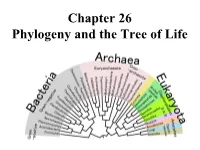
Chapter 26 Phylogeny and the Tree of Life • Biologists Estimate That There Are About 5 to 100 Million Species of Organisms Living on Earth Today
Chapter 26 Phylogeny and the Tree of Life • Biologists estimate that there are about 5 to 100 million species of organisms living on Earth today. • Evidence from morphological, biochemical, and gene sequence data suggests that all organisms on Earth are genetically related, and the genealogical relationships of living things can be represented by a vast evolutionary tree, the Tree of Life. • The Tree of Life then represents the phylogeny of organisms, the history of organismal lineages as they change through time. – In other words, phylogeny is the evolutionary history of a species or group of related species. • Phylogeny assumes that all life arise from a previous ancestors and that all organisms (bacteria, fungi, protist, plants, animals) are connected by the passage of genes along the branches of the phylogenetic tree. • The discipline of systematics classifies organisms and determines their evolutionary relationships. • Systematists use fossil, molecular, and genetic data to infer evolutionary relationships. • Hence, systematists depict evolutionary relationships among organisms as branching phylogenetic trees. • A phylogenetic tree represents a hypothesis about evolutionary relationships. • Taxonomy is the science of organizing, classifying and naming organisms. • Carolus Linnaeus was the scientist who came up with the two-part naming system (binomial system). – The first part of the name is the genus – The second part, called the specific epithet, is the species within the genus. – The first letter of the genus is capitalized, and the entire species name is italicized. • Homo sapiens or H. sapiens • All life are organize into the following taxonomic groups: domain, kingdom, phylum, class, order, family, genus, and species. (Darn Kids Playing Chess On Freeway Gets Squished). -

Answers to Common Misconceptions About Biological Evolution Leah M
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Papers in Evolution Papers in the Biological Sciences 5-2017 Answers to common misconceptions about biological evolution Leah M. Abebe University of Nebraska-Lincoln Blake Bartels University of Nebraska-Lincoln Kaitlyn M. Caron University of Nebraska-Lincoln Adam M. Gleeson University of Nebraska-Lincoln Samuel N. Johnson University of Nebraska-Lincoln See next page for additional authors Follow this and additional works at: http://digitalcommons.unl.edu/bioscievolution Part of the Evolution Commons Abebe, Leah M.; Bartels, Blake; Caron, Kaitlyn M.; Gleeson, Adam M.; Johnson, Samuel N.; Kluza, Tyler J.; Knopik, Nicholas W.; Kramer, Kristen N.; Maza, Masiel S.; Stava, Kaitlyn A.; Sullivan, Kaitlyn; Trimble, Jordan T.; and Zink, Robert M. , editor, "Answers to common misconceptions about biological evolution" (2017). Papers in Evolution. 4. http://digitalcommons.unl.edu/bioscievolution/4 This Article is brought to you for free and open access by the Papers in the Biological Sciences at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Papers in Evolution by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Authors Leah M. Abebe; Blake Bartels; Kaitlyn M. Caron; Adam M. Gleeson; Samuel N. Johnson; Tyler J. Kluza; Nicholas W. Knopik; Kristen N. Kramer; Masiel S. Maza; Kaitlyn A. Stava; Kaitlyn Sullivan; Jordan T. Trimble; and Robert M. Zink , editor This article is available at DigitalCommons@University of Nebraska - Lincoln: http://digitalcommons.unl.edu/bioscievolution/4 Answers to common misconceptions about biological evolution Class of BIOS 472, University of Nebraska, Lincoln, Spring 2017 Students: Leah M. Abebe, Blake Bartels, Kaitlyn M.