DNA Barcoding of Marine Metazoa
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Conservation of Handfish and Their Habitats – Annual Report Tim Lynch, Tyson Bessell, Alexander Hormann, Carlie Devine and Neville Barrett
Conservation of handfish and their habitats – annual report Tim Lynch, Tyson Bessell, Alexander Hormann, Carlie Devine and Neville Barrett Project A10 – Conservation of spotted handfish 28 February 2019 Milestone 4– Research Plan 4 (2018) www.nespmarine.edu.au Enquiries should be addressed to: Dr Tim P. Lynch Senior Research Scientist CSIRO Castray Esplanade [email protected] Project Leader’s Distribution List Derwent Estuary Program Ursula Taylor Zoo and Aquarium Association (ZAA) Craig Thorburn Natural Resource Management (NRM) Nepelle Crane South MAST Ian Ross Royal Yacht Club of Tasmania Nick Hutton Derwent Sailing Squadron Shaun Tiedemann The Handfish Recovery Team (HRT) See list below Marine and Freshwater Species Conservation Section Wildlife, Heritage and Marine Division Department of the Environment and Energy (DoEE) Threatened Species Policy and Andrew Crane Conservation Advice Branch Department of Primary Industries, Parks, Water and Environment (DPIPWE) Office of the Threatened Species Commissioner (DoEE) The project will also report its findings on a semi-annual basis to the National Handfish Recovery Team (NHRT) – see below. This is a governance body that is constituted between the Tasmanian State and the Commonwealth government with other interested parties: Department of the Environment and Energy (Commonwealth) Department of Primary Industries, Parks, Water and Andrew Crane Environment (Tas) CSIRO scientist, running current surveys and substrate trials Tim Lynch (Chair) University of Tasmania, handfish research Neville -
Appendicularia of CTAW
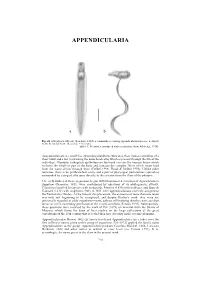
APPENDICULARIA APPENDICULARIA a b Fig. 26. Oikopleura albicans (Leuckart, 1854), a commonly occurring appendicularian species: a, dorsal view; b, lateral view. (Scale bar = 0.1 mm). [after T. Prentiss, reproduced with permission, from Alldredge 1976] Appendicularians are small free swimming planktonic tunicates, their bodies consisting of a short trunk and a tail (containing the notochord cells) which is present through the life of the individual. Glandular (oikoplast) epithelium on the trunk secretes the mucous house which encloses the whole or part of the body and contains the complex filters which strain food from the water driven through them (Deibel 1998; Flood & Deibel 1998). Unlike other tunicates, there is no peribranchial cavity and a pair of pharyngeal perforations (spiracles) surrounded by a ring of cilia open directly to the exterior from the floor of the pharynx. The early studies of these organisms, begun with Chamisso's description of Appendicularia flagellum Chamisso, 1821, were confounded by questions of its phylogenetic affinity. Chamisso classified his species with medusoids, Mertens (1830) with molluscs, and Quoy & Gaimard (1833) with zoophytes. Only in 1851 were appendicularians correctly assigned to the Tunicata by Huxley. At the time of this placement, the existence of more than one taxon was only just beginning to be recognised, and despite Huxley's work, they were not universally regarded as adult organisms—some authors still insisting that they were ascidian larvae or a free swimming generation of the sessile ascidians (Fenaux 1993). Subsequently, these questions were resolved by the work of Fol (1872) on material from the Straits of Messina, which forms the basis of later studies on the large collections of the great expeditions of the 19th century that revealed their true diversity in the oceanic plankton. -

Eurochordata and Cephalochordata of Persian Gulf & Oman See
Subphylum Urochordata : Tunicates Class Ascidacea Class Larvacea Class Thaliacea Subphylum Cephalochordata : Amphioxus Phylum Chordata : 1 - Subphylum Urochordata 2 Subphylum Cephalochordata (Subphylum Urochordata : Tunicates ) Class Ascidiacea Class Larvacea Class Thaliacea (Subphylum Cephalochordata ) Class Ascidiacea Ascidia Kowalevsky Patricia Kott D. lane David George & Gordon Paterson Herdmania momus savigny Herdmania momus kiamanensis Didemnum candidum savigny Phallusia nigra savigny Styela canopus savigny Class Ascidiacea : Order : Aplousobranchia Family : Polyclinidae : Aplidium cf. rubripunctum C.Monniot & F. Monniot 1997 Polyclinum constellatum Savigny, 1816 Family Didemnidae Didemnum candidum Savigny, 1816 Didemnum cf. granulatum Tokioka, 1954 Didemnum obscurum F. Monniot 1969 Didemnum perlucidum F. Monniot, 1983 Didemnum sp. Didemnum yolky C.Monniot & F. Monniot 1997 Diplosoma listerianum Milne Edwards, 1841 Order : Phlebobranchia Family : Ascidiidae Ascidia sp. Phallusia julinea Sluiter, 1915 Phallusia nigra Savigny, 1816 Order : Stolidobranchia Family : Styelidae Botryllus gregalis Sluiter, 1898 Botryllus niger Herdman, 1886 Botryllus sp. Eusynstyela cf. hartmeyeri Michaelsen, 1904 Polyandrocarpa sp. Styela canopus Savigny, 1816 Symplegma bahraini C.Monniot & F. Monniot 1997 Symplegma brakenhielmi Michaelsen, 1904 Family pyuridae Herdmania momus Savigny, 1816 Pyura curvigona Tokioka, 1950 Pyurid sp. Class Larvacea Larva Seymour Sewell Phylum Chordata Subphylum Urochordata * Class Larvacea Order Copelata Family Fritillariidae -

The Origins of Chordate Larvae Donald I Williamson* Marine Biology, University of Liverpool, Liverpool L69 7ZB, United Kingdom
lopmen ve ta e l B Williamson, Cell Dev Biol 2012, 1:1 D io & l l o l g DOI: 10.4172/2168-9296.1000101 e y C Cell & Developmental Biology ISSN: 2168-9296 Research Article Open Access The Origins of Chordate Larvae Donald I Williamson* Marine Biology, University of Liverpool, Liverpool L69 7ZB, United Kingdom Abstract The larval transfer hypothesis states that larvae originated as adults in other taxa and their genomes were transferred by hybridization. It contests the view that larvae and corresponding adults evolved from common ancestors. The present paper reviews the life histories of chordates, and it interprets them in terms of the larval transfer hypothesis. It is the first paper to apply the hypothesis to craniates. I claim that the larvae of tunicates were acquired from adult larvaceans, the larvae of lampreys from adult cephalochordates, the larvae of lungfishes from adult craniate tadpoles, and the larvae of ray-finned fishes from other ray-finned fishes in different families. The occurrence of larvae in some fishes and their absence in others is correlated with reproductive behavior. Adult amphibians evolved from adult fishes, but larval amphibians did not evolve from either adult or larval fishes. I submit that [1] early amphibians had no larvae and that several families of urodeles and one subfamily of anurans have retained direct development, [2] the tadpole larvae of anurans and urodeles were acquired separately from different Mesozoic adult tadpoles, and [3] the post-tadpole larvae of salamanders were acquired from adults of other urodeles. Reptiles, birds and mammals probably evolved from amphibians that never acquired larvae. -
Annelida, Amphinomidae) in the Mediterranean Sea with an Updated Revision of the Alien Mediterranean Amphinomids
A peer-reviewed open-access journal ZooKeys 337: 19–33 (2013)On the occurrence of the firewormEurythoe complanata complex... 19 doi: 10.3897/zookeys.337.5811 RESEARCH ARTICLE www.zookeys.org Launched to accelerate biodiversity research On the occurrence of the fireworm Eurythoe complanata complex (Annelida, Amphinomidae) in the Mediterranean Sea with an updated revision of the alien Mediterranean amphinomids Andrés Arias1, Rômulo Barroso2,3, Nuria Anadón1, Paulo C. Paiva4 1 Departamento de Biología de Organismos y Sistemas (Zoología), Universidad de Oviedo, Oviedo 33071, Spain 2 Pontifícia Universidade Católica do Rio de Janeiro , Rio de Janeiro, Brazil 3 Museu de Zoologia da Unicamp, Campinas, SP, Brazil 4 Departamento de Zoologia, Instituto de Biologia, Universidade Federal do Rio de Janeiro (UFRJ) , Rio de Janeiro, RJ, Brasil Corresponding author: Andrés Arias ([email protected]) Academic editor: C. Glasby | Received 17 June 2013 | Accepted 19 September 2013 | Published 30 September 2013 Citation: Arias A, Barroso R, Anadón N, Paiva PC (2013) On the occurrence of the fireworm Eurythoe complanata complex (Annelida, Amphinomidae) in the Mediterranean Sea with an updated revision of the alien Mediterranean amphinomids. ZooKeys 337: 19–33. doi: 10.3897/zookeys.337.5811 Abstract The presence of two species within the Eurythoe complanata complex in the Mediterranean Sea is reported, as well as their geographical distributions. One species, Eurythoe laevisetis, occurs in the eastern and cen- tral Mediterranean, likely constituting the first historical introduction to the Mediterranean Sea and the other, Eurythoe complanata, in both eastern and Levantine basins. Brief notes on their taxonomy are also provided and their potential pathways for introduction to the Mediterranean are discussed. -

SCAMIT Newsletter Vol. 1 No. 2 1982
TAXONOMIC INTERCAL1BRAT10N NEWSLETTER May, 1932 Number 2 Next Scheduled Meeting: June lA, 1982 at 9:30 a.m. Place: Marine Biological Consultants 9^7 Newhal1 Street Costa Mesa, California 92627 Topic Taxonomic Group: Amphinomidae, Euphrosinidae, Phyllodocidae MINUTES FROM MAY 17, 1982 Name Fourteen different names with accompanying acronyms were suggested. Because there were so many, it was decided to vote for a name by ballot and talley the results at the June meeting. One ballot is enclosed in this Newsletter. Please bring it to the next meeting or mail it to Ann Martin by June 11. Make additional copies of the ballot for each member of your Company. Dues Unfortunately dues are going to be ch-arged. The dues will help defray costs incurred for storing the reference collection, paper, postage, and other unforeseen expenses. Dues will be $5.00 per year beginning June, 1982. This amount will be adjusted after six months if it is insufficient. Dues will be collected at the June meeting. For those unable to attend the meeting, dues can be mailed to.Ann Martin and a receipt will be mailed back. Specimen Exchange. Several points concerning the specimen exchange were brought out that will help. - Participants should be responsible for getting their specimens to the meeting, either in person or by mail. - Mark the specimens sequentially to prevent mixing of different groups. - Bring the voucher specimens to meeting to check them with ones from the specimen exchange. - When deciding what species to select for the specimen exchange, first call Tony Phillips, (213) 322-3131 extension 269, and tell him what you plan to bring in. -

Deepseacorals.Pdf
Protection of Deep-Sea Corals from Physical Damage by Fishing Gear under the MSA Deep Sea Coral Discretionary Authority Purpose The National Oceanic and Atmospheric Administration (NOAA) is a steward of the nation’s living marine resources. This document will assist NOAA offices and the regional fishery management councils (Councils)1 when developing protective measures for deep-sea corals under section 303(b)(2)(B) of the Magnuson-Stevens Fishery Conservation and Management Act (MSA).2 Section 303(b)(2) provides that any fishery management plan (FMP) which is prepared by any Council or the Secretary, with respect to any fishery, may: A) designate zones where, and periods when, fishing shall be limited, or shall not be permitted, or shall be permitted only by specified types of fishing vessels or with specified types and quantities of fishing gear; B) designate such zones in areas where deep sea corals are identified under section 408 [the Deep Sea Coral Research and Technology Program], to protect deep sea corals from physical damage from fishing gear or to prevent loss or damage to such fishing gear from interactions with deep sea corals, after considering long-term sustainable uses of fishery resources in such areas. 16 U.S.C. § 1853(b)(2)(A)-(B). We encourage use of this discretionary authority to advance the agency’s and Councils’ conservation objectives. NOAA’s Strategic Plan for Deep-Sea Coral and Sponge Ecosystems seeks to ensure that fisheries that may interact with known and likely deep-sea coral ecosystems are identified and monitored and that such ecosystems are protected from the impacts of fishing gear (see Figure 1).3 This document is consistent with those policy goals. -

Cirripedia of Madeira
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Universidade do Algarve Helgol Mar Res (2006) 60: 207–212 DOI 10.1007/s10152-006-0036-5 ORIGINAL ARTICLE Peter Wirtz Æ Ricardo Arau´jo Æ Alan J. Southward Cirripedia of Madeira Received: 13 September 2005 / Revised: 12 January 2006 / Accepted: 13 January 2006 / Published online: 3 February 2006 Ó Springer-Verlag and AWI 2006 Abstract We give a list of Cirripedia from Madeira phers. The marine invertebrates have been less studied Island and nearby deep water, based on specimens in and there has been no compilation of cirripede records the collection of the Museu Municipal do Funchal for Madeira, comparable to those for the Azores (Histo´ria Natural) (MMF), records mentioned in the archipelago (Young 1998a; Southward 1999). We here literature, and recent collections. Tesseropora atlantica summarize records from Madeira and nearby deep water Newman and Ross, 1976 is recorded from Madeira for and discuss their biogeographical implications. the first time. The Megabalanus of Madeira is M. az- oricus. There are 20 genera containing 27 species, of which 22 occur in depths less than 200 m. Of these Methods shallow water species, eight are wide-ranging oceanic forms that attach to other organisms or to floating The records are based on (1) the work of R.T. Lowe, objects, leaving just 13 truly benthic shallow water who sent specimens to Charles Darwin; (2) material in barnacles. This low diversity is probably a consequence the Museu Municipal do Funchal (Histo´ria Natural) of the distance from the continental coasts and the (MMF); (3) casual collecting carried out by residents or small area of the available habitat. -

Metabolomic Profiling Reveals Deep Chemical Divergence Between Two
OPEN Metabolomic profiling reveals deep SUBJECT AREAS: chemical divergence between two METABOLOMICS CHEMICAL ECOLOGY morphotypes of the zoanthid BIODIVERSITY MASS SPECTROMETRY Parazoanthus axinellae Nadja Cachet1, Gre´gory Genta-Jouve1,2, Julijana Ivanisevic1,3, Pierre Chevaldonne´3, Fre´de´ric Sinniger4,5, Received Ge´rald Culioli1,6, Thierry Pe´rez3 & Olivier P. Thomas1,3 10 October 2014 Accepted 1Institut de Chimie de Nice - EEIC, UMR 7272 CNRS, Universite´ de Nice-Sophia Antipolis, Parc Valrose, 06108 Nice, France, 8 January 2015 2Laboratoire de Pharmacognosie et de Chimie des Substances Naturelles, UMR CNRS 8638 COMETE, Universite´ Paris Descartes, 4 Avenue de l’Observatoire 75006 Paris, France, 3Institut Me´diterrane´en de Biodiversite´ et d’Ecologie Marine et Continentale, UMR Published 7263 CNRS, IRD, Aix Marseille Universite´, Avignon Universite´, Station Marine d’Endoume, Rue Batterie des Lions, 13007 6 February 2015 Marseille, France, 4Japan Agency for Marine-Earth Science and Technology, 224-3 Aza-Toyohara, Nago City, Okinawa 905- 2172, Japan, 5Tropical Biosphere Reseach Center, University of the Ryukyus, 3422 Sesoko, Motobu, Okinawa 905-0227, Japan, 6MAPIEM, EA 4323 Universite´ de Toulon, 83957 La Garde, France. Correspondence and requests for materials Metabolomics has recently proven its usefulness as complementary tool to traditional morphological and should be addressed to genetic analyses for the classification of marine invertebrates. Among the metabolite-rich cnidarian order T.P. (thierry.perez@ Zoantharia, Parazoanthus is a polyphyletic genus whose systematics and phylogeny remain controversial. imbe.fr) or O.P.T. Within this genus, one of the most studied species, Parazoanthus axinellae is prominent in rocky shallow (olivier.thomas@unice. waters of the Mediterranean Sea and the NE Atlantic Ocean. -

Conserving Critically Endangered Spotted Handfish
Conserving Critically Endangered spotted handfish Unique and quirky, spotted handfish (Brachionichthys hirsutus) are recognisable by their modified fins that resemble human hands. Once common in southern Tasmania’s Derwent estuary, spotted handfish experienced a severe decline in the 1980s. In 1996 they became the first marine fish to be listed as Critically Endangered by the IUCN Red List of Threatened Species. They are also listed as Critically Endangered under the Commonwealth Environment Protection and Biodiversity Conservation Act 1999, and Endangered under Tasmania's Threatened Species Protection Act 1995. Spotted handfish were once common in Tasmania’s Derwent estuary, but their populations experienced CSIRO, University of Tasmania (UTAS), the a serious decline in the 1980s. Tasmanian and Australian governments and the Derwent Estuary Program (DEP) have between 5–15m. Within bays they occupy been working together to conserve spotted habitats with more complex features such as handfish since the mid-1990s. depressions in the seabed made by stingrays, or fields of sea-squirts. Distribution Lacking swim bladders, spotted handfish use Handfish belong to a group of coastal their modified fins to ‘walk’ across the seabed anglerfish with a narrow distribution in south- rather than swim. Movement studies suggest eastern Australia. There are 14 species with they only travel small distances: 10m–460m seven endemic to Tasmania and Bass Strait. over many months, or an average of 4m a day. Spotted handfish were once prevalent along Spotted handfish are ambush predators and, Tasmania’s eastern coast, and were so like their close cousins the deep-sea angler common that during the 1960s and ‘70s that fishes, they have a lure located just above the they were routinely collected for practical mouth, perhaps to entice their prey of demonstrations at Hobart’s university. -

Barnacle Editor Workshop
Barnacle Editor Workshop VLIZ InnovOcean Site Wandelaarkaai 7 – entrance Pakhuis 61 (UNESCO) B-8400 Oostende, Belgium February 24-28, 2020 Final Report Barnacle Participants: Keith Crandall, Meeting Organizer, George Washington University, Washington, DC USA Jens Hoeg, University of Copenhagen, Copenhagen, Denmark Marcos Pérez-Losada, George Washington University, Washington, DC USA Benny Chan, Academia Sinica, Taipei, Taiwan Henrick Glenner, University of Bergen, Bergen, Norway Andy Gale, University of Portsmouth, Portsmouth, United Kingdom Niklas Dreyer, Academia Sinica, Taipei, Taiwan WoRMS Data Management Team (DMT): Stefanie Dekeyzer (Meeting Coordinator) Bart Vanhoorne Wim Decock Leen Vandepitte Target Group: The barnacles – more specifically, the broader group of Thecostraca including the traditional barnacles (Cirripedia) as well as the related groups of Facetotecta and Ascothoracida. The thecostracan barnacles rank among the most commonly encountered marine crustaceans in the world. They deviate from almost all other Crustacea in that only the larvae are free-living, while the adults are permanently sessile and morphologically highly specialized as filter feeders or parasites. In the most recent classifications of the crustacean Maxillopoda 1 and latest phylogenetic analyses 2-4 the Thecostraca sensu Grygier 5, comprising the Facetotecta, Ascothoracida, and Cirripedia, form monophyletic assemblages. Barnacle phylogenetics has advanced greatly over the last 10 years. Nonetheless, the relationships and taxonomic status of some groups within these three infraclasses are still a matter of debate. While the barnacles where the focus of Darwin’s detailed taxonomic work, there has not been a comprehensive review of the species of barnacles as a whole since Darwin. As a consequence, the barnacle entries within the WoRMS Database is woefully out of date taxonomically and missing many, many species and higher taxa. -

Handfish Survey in the Vicinity of a Proposed Intake Pipeline
HANDFISH SURVEY IN THE VICINITY OF A PROPOSED INTAKE PIPELINE AND EXISTING INTAKE INSPECTION AT CRAYFISH POINT, TAROONA, DERWENT ESTUARY Report to IMAS March 2018 www.marinesolutions.net.au © Marine Solutions 2018. This document should only be used for the specific project and purposes for which it was commissioned. 1 Version Author Date Reviewed by reviewed 1 of 1 Annie Ford and 20/03/2018 Laura Smith Joanna Smart Note: Location maps throughout this report are representative only; for precise GPS coordinates, see the appendices. All satellite imagery used throughout is sourced from The Land Information System Tasmania (LIST). 1 Cover photo, IMAS Taroona, November 2016 (photo by Marine Solutions). Handfish survey at site of proposed intake pipeline, Taroona 2 TABLE OF CONTENTS Table of Contents .......................................................................................................................................... 3 Table of Figures ............................................................................................................................................. 5 1 Executive Summary ............................................................................................................................... 6 2 Introduction .......................................................................................................................................... 8 2.1 Background ..................................................................................................................................