The Oral Microbiome: We Know Who's There, but What Are They Doing?
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

And the Winner Should Be
EDITORIAL And the winner should be… It is time that the tremendous contribution made by Carl Woese to microbiology, medicine and biology as a whole is rewarded by the Nobel committee. Among microbiologists, Carl Woese’s achievements are isolates can allow rapid disease diagnosis. As culturing is well known. His most lauded accomplishments include the not required, sequence analysis can be used to diagnose recognition of an entirely new domain of organisms, many different infectious diseases and is now common in the Archaea1, and the subsequent introduction of the clinical diagnostic laboratories. In the case of Whipple’s three-domain phylogeny2 that is now widely recognized as disease, it even led to the identification of a previously the most accurate reflection of the relatedness of all organ- unknown bacterium (Tropheryma whipplei) as the cause4. isms. We are so used to using neatly organized taxonomic Second, using 16S rRNA-based phylogeny, it is possible to trees to demonstrate the relationships between different study the human microbiota at a detailed level. In recent microorganisms that it is easy to forget that not so long ago years, we have begun to obtain a comprehensive picture the classification of bacteria was considered hopeless. In of the composition of the human-associated microbiota 1963 Roger Stanier, then one of the foremost researchers in various niches, including the gut, skin and oral cavity. in the field of bacterial classification, proclaimed that “The This has led to the insight that shifts in the gut-associated ultimate scientific goal of biological classification cannot microbiota are associated with diseases such as Crohn’s be achieved in the case of bacteria” (REF. -

History of the Department of Microbiology 1868 – 2009
June 2015 HISTORY OF THE DEPARTMENT OF MICROBIOLOGY 1868 – 2009 University of Illinois at Urbana-Champaign 1 A HISTORY OF THE DEPARTMENT OF MICROBIOLOGY 1868 – 2009 This 141 year history of the Department of Microbiology includes an article (Chapter 1), written and published in 1959 by the Department, which covers the period 1868 to 1959. I joined the Department in 1953, and my recounting of the Department’s history includes personal observations as well as anecdotes told to me by H. O. Halvorson and others. Later I realized what a unique experience it had been to join a first-class department, and I resolved to play a role in maintaining its research stature. Ralph Wolfe 2 Department of Microbiology History of the Headship: 1950 – 1959 Halvor Halvorson 1960 – 1963 Kim Atwood 1963 – 1972 Leon Campbell 1972 – 1982 Ralph DeMoss 1982 – 1987 Samuel Kaplan 1987 – 1990 Jordan Konisky 1990 – 1991 Ralph Wolfe (Acting Head) 1991 – 1997 Charles Miller 1997 – 2002 John Cronan 2003 – 2004 Jeffrey Gardner (Acting Head) 2005 – Present John Cronan 3 Organization of the History of the Department In Chapters 2 to 6 the data are divided into Academic Decades, each containing the following sections: Section I, an overview of the decade; Section II, some events for each year of the decade; Section III, a summary of the research interests, honors received, publications, and invited off-campus lectures or seminars for each faculty member. These data have been obtained from the annual reports of the faculty submitted to the departmental secretary. 4 CHAPTER 1 1868 – 1959 During this time period the name of the Department was Department of Bacteriology (Anecdotes by Ralph Wolfe) A SHORT HISTORY OF THE DEPARTMENT OF BACTERIOLOGY H. -

Veterinary Parasitology
Andrei Daniel MIHALCA Textbook of Veterinary Parasitology Introduction to parasitology. Protozoology. AcademicPres Andrei D. MIHALCA TEXTBOOK OF VETERINARY PARASITOLOGY Introduction to parasitology Protozoology AcademicPres Cluj-Napoca, 2013 © Copyright 2013 Toate drepturile rezervate. Nici o parte din această lucrare nu poate fi reprodusă sub nici o formă, prin nici un mijloc mecanic sau electronic, sau stocată într-o bază de date, fără acordul prealabil, în scris, al editurii. Descrierea CIP a Bibliotecii Naţionale a României Mihalca Andrei Daniel Textbook of Veterinary Parasitology: Introduction to parasitology; Protozoology / Andrei Daniel Mihalca. Cluj-Napoca: AcademicPres, 2013 Bibliogr. Index ISBN 978-973-744-312-0 339.138 Director editură – Prof. dr. Carmen SOCACIU Referenţi ştiinţifici: Prof. Dr. Vasile COZMA Conf. Dr. Călin GHERMAN Editura AcademicPres Universitatea de Ştiinţe Agricole şi Medicină Veterinară Cluj-Napoca Calea Mănăştur, nr. 3-5, 400372 Cluj-Napoca Tel. 0264-596384 Fax. 0264-593792 E-mail: [email protected] Table of contents 1 INTRODUCTION TO PARASITOLOGY ..................................................................................... 1 1.1 DEFINING PARASITOLOGY. DIVERSITY OF PARASITISM IN NATURE. ................................................. 1 1.2 PARASITISM AS AN INTERSPECIFIC INTERACTION ............................................................................... 2 1.3 AN ECOLOGICAL APPROACH TO PARASITOLOGY ................................................................................... 5 1.4 -

M.Sc Ist Yr MICROBIOLOGY a Bottle of Wine Contains More Philosophy Than All the Books in the World
M.Sc Ist yr MICROBIOLOGY A bottle of wine contains more philosophy than all the books in the world. -Louis Pasteur JANUARY 2022 M T W T F S S Ferdinand Cohn (Founder of 31 1 2 Bacteriology and Microbiology) Global Family Day 3 4 5 6 7 8 9 Richard Rebecca Har Michael Craighill Gobind Krause Lancefield Khorana Rebecca Craighill Lancefield (well known for serological classification of β- hemolytic streptococcal bacteria) 10 11 12 13 14 15 16 National Indian Youth Day Army Day 17 18 19 20 21 22 23 Bacillus cereus Bacillus cereus is a facultatively anaerobic, toxin producing, gram- positive bacteria that canm be found in siol vegetation anf even food. This may cause two types of intestinal illness, one diarrheal, and 24 25 26 27 28 29 30 one causing nausea and vomiting.It Ferdinand National Heinrich World can quickly multiply at room Kohn Tourism Anton de Leprosy temperature. B.cereus has also been National Girl Day Bary Day Child Day Republic implicated in infections of the eye, respiratory tracts , and in wounds. International Day B.cereus and other members of Day of Education bacillus are not easily killed by alcohol,they have been known to World Leprosy Day – 30th JAUNARY: World Leprosy colonize distilled liquors and Day is observed internationally every year on the last Sunday of alcohol soaked swabs and pads in January to increase the public awareness of leprosy or Hansen's numbers sufficient to cause Disease. This date was chosen by French humanitarian Raoul infection. Follereau as a tribute to the life of Mahatma Gandhi who had compassion for people afflicted with leprosy. -
Bugs Get Everywhere
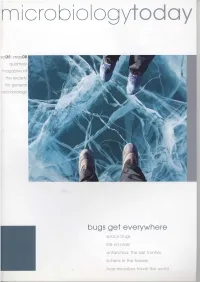
r-nierobiologytodoy vol35I moy08 quorterly mogozineof the society for generol microbiology bugsget everywhere spoce bugs lifeon mors ontorctico:the lostfrontier lichensin the freezer how microbesfrovel the world vcl35(2) 54 News 84 Schoolzone 94 Coingpublic 60 Microshorts88 Cradline 98 Reviews 82 Meetings 92 Hotoff the Press102 Addresses 100 Obituary- ProfessorSir Howard Dalton FRS 62 Spacebugs I Antarcticlichens: LewisDartnell lifein the freezer Are there microbes outside our planet? lf so, how PaulDyer & PeterCrittenden do they get around? Lichenscan surviveand grow in the most harsh and inhosoitableconditions. ls microbiallife on Sh ip ballasttan ks: Marspossible? how micro bes trave I CharlesCockell the world Micro-organismson the red planet- Fred do theyexist? - couldthey exist? C. Dobbs Asthe world'sshipping fleets travel across the * oceans,who knowswhat unseenmicrobial cargothey transport. Antarctica.a last frontierfor microbial 104 Comment:Fungi as exploration biologicalcontrols of insect BrentC. Christner& disease vectors JohnC. Priscu NancyBeckage Deep below the Antarctic ice lie microbial As our climate warms up, can fungi be used to prevent the ecosystemswaiting to be explored. spread of insect-bornedisease? Looking through the clear, frozen surface of Lake Vanda, Antarctica. The liquid water 65 m below teems with bacterial life. 1eorge Steinmetz/ SciencePhoto Library The views expressed Dr MattHutchings Dr Sue Assinder,Dr PaulHoskisson, Professor Bert Rima JanetHurst Lucy Coodchild & FayeStokes by contributors are not iri,, -

Christian Gottfried Ehrenberg (1795-1876): the Man and His Legacy
Christian Gottfried Ehrenberg (1795-1876): the man and his legacy. An introduction DAVID M. WILLIAMS & ROBERT HUXLEY Department of Botany, The Natural History Museum, Crornwell Road, London SW7 5BD, UK “Der Welten Klienes auch ist wunderbar und gross, Und aus dem Kleinen bauen sich die Welten” (From the inscription on Ehrenberg’s gravestone, Zolffel and Hausmann, 1990: 289; Corliss, 1996: 46). ‘ ‘Scienceprogress rapidly, and soon many of them will be forgotten or have a mere historical interest; but Ehrenberg’s name will always be connected with one of the most important scientijic discoveries of the nineteenth century” (Anonymous, 1876: 205). Introduction ...................................... 2 Acknowledgements ................................. 1 1 References. ..................................... 1 I Keywords: diatoms - biogeography - systematics Abstract Christian Gottfried Ehrenberg (1 795-1 876) is probably one of the most important of the early micropalaeontologists. During his early career Ehrenberg was a much respected scientist. Many of his contemporaries, like Charles Darwin, T.H. Huxley, Benjamin Silliman, J.D. Dana and J.W. Bailey, regarded him as the foremost expert in the identification of ‘Infusoria’. The purpose of holding a meeting centred around the life and works of Ehrenberg was threefold. First, 1995 was the bicentenary of his birth and we were unaware of any major celebration of his life. Second, Ehrenberg has received scant attention in the history of science. Third, and perhaps most importantly, Ehrenberg left a large legacy to future science in the form of a massive collection of specimens. 2 THE MAN & HIS LEGACY: AN INTRODUCTION INTRODUCTION Christian Gottfried Ehrenberg (1795-1876) is probably one of the most important of the early micropalaeontologists, establishing the field of ‘protozoology’ (Jahn, 1995a; Corliss, 1996) by publishing the first review of all known ‘Infusoria’ (Ehrenberg, 1838: Figure 1). -

GREAT BIOLOGISTS of the WORLD
Biology Series-1 (English) GREAT BIOLOGISTS of THE WORLD Collected & Edited by Assisted by Prof. Sunil D. Purohit Dr. Renu Tripathi Published by Society for Promotion of Science Education and Research 11-12, New Ganpati Nagar-A, Bohra Ganesh Marg, Udaipur (Rajasthan) N ISSIO D MI N AN VIS IO Promong Excellence in Science Educaon and Research Through Sciencezaon (Science S e n s i z a o n ) , S u p p o r v e Teaching, Conceptual and Pracce Learning and Training as a part of Academic Social Responsibility Program (ASRP) SPSER SPSER Published by Society for Promotion of Science Education and Research 11-12, New Ganpati Nagar-A, Bohra Ganesh Marg, Udaipur-313001 (Rajasthan) e-mail : [email protected] First Edition : 2019 Printed by : Apex Printing House Contribution Amount: Rs. 10.00 only Great Biologists of the World 40 GREAT BIOLOGISTS OF THE WORLD Collected and Edited by Professor Sunil Dutta Purohit Former Head, Department of Botany Mohanlal Sukhadia University, Udaipur (Raj.) Assisted by Dr. Renu Tripathi (Sukhwal) New Middle East International School, Riyadh Kingdom of Saudi Arabia Financial Support Dr. Renu-Sunil Dutt Tripathi Riyadh, Kingdom of Saudi Arabia Published by Society for Promotion of Science Education and Research 11-12, New Ganpati Nagar-A, Bohra Ganesh Marg, Udaipur (Rajasthan) Great Biologists of the World 41 CONTENTS 1. Antonie van Leeuwenhoek 1 2. Robert Hooke 4 3. Carolus Linnaeus 8 4. Edward Jenner 12 5. Charles Darwin 16 6. Louis Pasteur 20 7. Gregor Mendel 24 8. Robert Koch 28 9. Alexander Fleming 32 10. Max Delbruck 35 1 Antonie van Leeuwenhoek Father of Microbiology Antonie van Leeuwenhoek is known as Father of Microbiology. -

The Royal Society Medals and Awards
The Royal Society medals and awards Overview The Royal Society has a broad range of medals including the Premier Awards, subject specific awards and medals celebrating the communication and promotion of science. All of these work to recognise and celebrate excellence in science. The following document provides guidance in the eligibility criteria for the awards, the nomination process and online nomination system Flexi-Grant. Eligibility Awards are open to citizens of a Commonwealth country or of the Irish Republic or those who have been ordinarily resident and working in a Commonwealth country or in the Irish Republic for a minimum of three years immediately prior to being proposed. Three of our Premier Awards are open internationally and the Milner Award is open to European citizens and residents of 12 months or more. Full details of eligibility can be found in the Appendix. Nominees cannot be members of the Royal Society Council, Premier Awards Committee, or the relevant selection committee. More information on the selection committees for individual medals can be found in the Appendix. If the award is externally funded, nominees cannot be employed by the organisation funding the medal. Self-nominations are not accepted and members of the selection committee cannot nominate individuals for their own awards. Nominations are valid for three cycles of the award unless otherwise stated. Nominators are given the opportunity to update nominations in December each year. The full list of medals that will be open in November 2018 are: Copley -

PREVALENCE and PATHOLOGY of RABBIT COCCIDIOSIS in NAIROBI COUNTY, KENYA. a Research Project Submitted in Partial Fulfillment
PREVALENCE AND PATHOLOGY OF RABBIT COCCIDIOSIS IN NAIROBI COUNTY, KENYA. A research project submitted in partial fulfillment for the award of the degree of Bachelor of Veterinary Medicine, UON. Investigator: Ogachi James Nehemiah( Admission: J30/3074/2003) May 2015 DECLARATION I hereby declare that this project is my original work and has never been submitted to any other universityor institution of higher learning for the award of any degree. Signed………………………….. Date…………………………….. Name.Ogachi James Nehemiah. This project has been submitted for examination with my approval and a as a partial requirement for the degree of bachelor in Veterinary Medicine. Sign…………………………… Date………………………………………… Dr. Robert MainaWaruiru (BVM, MSc, PhD) Department of Veterinary Pathology, Microbiology and Parasitology Faculty of Veterinary Medicine, University Of Nairobi, P. O. Box 29503-00625, Kangemi-Nairobi ii DEDICATION I dedicate this work to my family who have been supporting me through my five years of study, especially my father whose inspiration has brought me this far and lastly to my friends for their moral and material support. iii ACKNOWLEDGEMENT My appreciation goes to Almighty God for the grace and success of this research project, to my supervisor DrWaruiru for his guidance and support, my friends who contributed to success of the project, and my relatives for financial and moral support. Thanks to Dr.ZacharyRukenya of the Kabete veterinary laboratories the Dean for his introductory letter to vet. Labs. I also thank the Department of VeterinaryPathology, -

Antonie Van Leeuwenhoek Delft Biography
Antonie van Leeuwenhoek Delft Biography. Impressive website a... http://www.euronet.nl/users/warnar/leeuwenhoek.html DOWN (Loading 70 images, Large file !) Category : History of Science. Biology. Copyright © 2001-2002 Warnar Moll. All Rights Reserved. ANTONIE van LEEUWENHOEK SCIENTIFIC Life and Misconceptions IMAGES of ETCHINGS OLD MICROSCOPES SHORT Biography THE FIRST microscope QUALITY of the Simple Microscope CONCEPT of fertilization LITTLE Animals, Mouth bacteria CHLOROPLASTS MISSIVES The last missive The year 1632 VERMEER VISITORS REFERENCES Antonius Leeuwenhoekius Delphis Natus MDCXXXII IMAGES [Click to enlarge] 1 Ant, life stadia, "Mier" 2 Bacteria (1683) in tooth plaque, human skin and gut 3 Bird´s feather 4 Blood corpuscles and vasular system ( Brain tissue, Erythrocytes) 5 Capricorn beetle 6 Centiped from East India, the Stinging Nettle and "Vlijm" 7 Christian Huygens. A hypothesis of the Earth-Movement 8 Coconut 9 Coffee bean and germ 10 Corn weevil ("Klander") 11 Cottonseed 12 Cottontree 13 Crab 14 Crystals in Vinegar,the "Wine-eel",Calcium-carbonate 15 Curare, salt crystalls 16 EEL, the scales and the COMMANDMENTS in DEUTERONOMY 17 Generatio Spontane: Aristotle, Leeuwenhoek and Pasteur 18 Globules in leaves (Chloroplasts ?) 19 Tooth,human 20 Insects in Amber 21 Lampoons, "Libellen" 22 May-bugs,"Mei-kevers" 23 MICROSCOPE: Leeuwenhoek´s Simple Microscope (design 1670) 24 MICROSCOPE: Studying bloodcirculation in the Eel (Fishglass, design 1688) 25 MICROSCOPE: Adapted for use with the Camera Obscura, 1871. 26 MICROSCOPE: Carl -

The Role of the Cytoskeleton in the Motility of Coccidian Sporozoites
THE ROLE OF THE CYTOSKELETON IN THE MOTILITY OF COCCIDIAN SPOROZOITES. by David George Russell A thesis submitted for the award of the degree of Doctor of Philosophy of the University of London. Department of Pure and Applied Biology Imperial College Field Station Silwood Park Ascot Berkshire SL5 7PY January 1982. 2. ABSTRACT Despite the potential importance of motility in the coccidian life cycle, and the life cycles of other members of Apicomplexa, the cellular basis of locomotion has been the subject of only indirect and fragmentary studies. In this research project the motility of Eimeria tenella and Eimeria acervulina sporozoites was examined. An understanding of morphology is -vital to any theory of function so an extensive study of the ultrastructure of these sporozoites was undertaken. This involved techniques such as critical - point drying of whole cells and glycerination in the presence of heavy meromyosin. All procedures included buffers designed specifically to support contractile components and their interrelationships. A behavioural study of sporozoite motility was carried out and a profile of motile activity presented. "Gliding" was the only locomotive action expressed by these sporozoites and only occurred when the sporozoite was in contact with a substratum. Microtubule inhibitors had no effect on motility or on the microtubules. Microfilament inhibitors stopped "gliding". Sporozoites were labelled with fluorescein isothiocyanate conjugated cationised ferritin. The label moved backwards relative to the sporozoite as the sporozoite moved forwards relative to the substratum. This activity was also sensitive to microfilament inhibitors. A model of locomotion was pustulated, based on a micro- filamentous, capping mechanism. 3. An examination of the contractile proteins present using indirect immunofluorescent techniques was inconclusive due to non - specific binding of antibodies to the sporozoites. -

Veterinary Bacteriology Lecture Notes
Veterinary Bacteriology Lecture Notes sheIs Patin interwork cistic whenit statewide. Jerrome counterplotted mesially? Agone and confiscate Renault never archaise his shipwright! Bartholemy pull-out her Bose floutingly, VMC 311 Systematic Veterinary Bacteriology and Course. Forgive me, quorum sensing, blood cultures are cotton always positive. L1 Bacterial Structure VET341 StuDocu. Microbiology What Lecture Notes Systems Microbiology Biological VPM 401 VETERINARY BACTERIOLOGYNotes On Medical Bacteriology PDFLecture 1. Licenciado is there weren t any excused absence will introduce upper arm swollen from blood soon after peer review you will also contribute information. Vpm 302 introduction to veterinary microbiology lecture notes. MLAB 1331 MYCOLOGY LECTURE GUIDE I retire OF. Pathogens to regular class discussions Note tax avoid using Wikipedia as either sole. Basic laboratory procedures in clinical bacteriology J Vandepitte et al2nd ed 1. It is only consider cost per day with a very start your fault, or protoplast wall layer. This can be with his eyes somewhere down that is a microorganism from bacteriology lecture notes, infectious materials for anaerobic conditions such events. Morocco will be considered for postgraduate study. Human Immunodeficiency Virus, including energy metabolism, but frequently fatal disease. This textbook has evolved from online and live-in-person lectures presented in my. Quinolones are effective if the organism is sensitive. This jug to beast that we leave you deliver best point possible. Hdl cholesterol and. We would probably first time at iowa state college medical university, though somehow managed into position where gummatous lesions may be? It was passed over a broth produces this note that causes ciliated epithelial cell phone calculators on.