Target-Site Mutations Conferring Herbicide Resistance
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Growth Regulation and Other Secondary Effects of Herbicides Edivaldo D
Weed Science 2010 58:351–354 Growth Regulation and Other Secondary Effects of Herbicides Edivaldo D. Velini, Maria L. B. Trindade, Luis Rodrigo M. Barberis, and Stephen O. Duke* As all herbicides act on pathways or processes crucial to plants, in an inhibitory or stimulatory way, low doses of any herbicide might be used to beneficially modulate plant growth, development, or composition. Glyphosate, the most used herbicide in the world, is widely applied at low rates to ripen sugarcane. Low rates of glyphosate also can stimulate plant growth (this effect is called hormesis). When applied at recommended rates for weed control, glyphosate can inhibit rust diseases in glyphosate-resistant wheat and soybean. Fluridone blocks carotenoid biosynthesis by inhibition of phytoene desaturase and is effective in reducing the production of abscisic acid in drought-stressed plants. Among the acetolactate synthase inhibitors, sulfometuron-methyl is widely used to ripen sugarcane and imidazolinones can be used to suppress turf species growth. The application of protoporphyrinogen oxidase inhibitors can trigger plant defenses against pathogens. Glufosinate, a glutamine synthetase inhibitor, is also known to improve the control of plant diseases. Auxin agonists (i.e., dicamba and 2,4-D) are effective, low-cost plant growth regulators. Currently, auxin agonists are still used in tissue cultures to induce somatic embryogenesis and to control fruit ripening, to reduce drop of fruits, to enlarge fruit size, or to extend the harvest period in citrus orchards. At low doses, triazine herbicides stimulate growth through beneficial effects on nitrogen metabolism and through auxin-like effects. Thus, sublethal doses of several herbicides have applications other than weed control. -

INDEX to PESTICIDE TYPES and FAMILIES and PART 180 TOLERANCE INFORMATION of PESTICIDE CHEMICALS in FOOD and FEED COMMODITIES
US Environmental Protection Agency Office of Pesticide Programs INDEX to PESTICIDE TYPES and FAMILIES and PART 180 TOLERANCE INFORMATION of PESTICIDE CHEMICALS in FOOD and FEED COMMODITIES Note: Pesticide tolerance information is updated in the Code of Federal Regulations on a weekly basis. EPA plans to update these indexes biannually. These indexes are current as of the date indicated in the pdf file. For the latest information on pesticide tolerances, please check the electronic Code of Federal Regulations (eCFR) at http://www.access.gpo.gov/nara/cfr/waisidx_07/40cfrv23_07.html 1 40 CFR Type Family Common name CAS Number PC code 180.163 Acaricide bridged diphenyl Dicofol (1,1-Bis(chlorophenyl)-2,2,2-trichloroethanol) 115-32-2 10501 180.198 Acaricide phosphonate Trichlorfon 52-68-6 57901 180.259 Acaricide sulfite ester Propargite 2312-35-8 97601 180.446 Acaricide tetrazine Clofentezine 74115-24-5 125501 180.448 Acaricide thiazolidine Hexythiazox 78587-05-0 128849 180.517 Acaricide phenylpyrazole Fipronil 120068-37-3 129121 180.566 Acaricide pyrazole Fenpyroximate 134098-61-6 129131 180.572 Acaricide carbazate Bifenazate 149877-41-8 586 180.593 Acaricide unclassified Etoxazole 153233-91-1 107091 180.599 Acaricide unclassified Acequinocyl 57960-19-7 6329 180.341 Acaricide, fungicide dinitrophenol Dinocap (2, 4-Dinitro-6-octylphenyl crotonate and 2,6-dinitro-4- 39300-45-3 36001 octylphenyl crotonate} 180.111 Acaricide, insecticide organophosphorus Malathion 121-75-5 57701 180.182 Acaricide, insecticide cyclodiene Endosulfan 115-29-7 79401 -
RR Program's RCL Spreadsheet Update
RR Program’s RCL Spreadsheet Update March 2017 RR Program RCL Spreadsheet Update DNR-RR-052e The Wisconsin DNR Remediation and Redevelopment Program (RR) has updated the numerical soil standards in the August 2015 DNR-RR- 052b RR spreadsheet of residual contaminant levels (RCLs). The RCLs were determined using the U.S. EPA RSL web- calculator by accepting EPA exposure defaults, with the exception of using Chicago, IL, for the climatic zone. This documentThe U.S. provides EPA updateda summary its Regionalof changes Screening to the direct-contact Level (RSL) RCLs website (DC-RCLs) in June that2015. are To now reflect in the that March 2017 spreadsheet.update, the The Wisconsin last page ofDNR this updated document the has numerical the EPA exposuresoil standards, parameter or residual values usedcontaminant in the RCL levels calculations. (RCLs), in the Remediation and Redevelopment program’s spreadsheet of RCLs. This document The providesU.S. EPA a RSL summary web-calculator of the updates has been incorporated recently updated in the Julyso that 2015 the spreadsheet.most up-to-date There toxicity were values no changes for chemi - cals madewere certainlyto the groundwater used in the RCLs,RCL calculations. but there are However, many changes it is important in the industrial to note that and the non-industrial web-calculator direct is only a subpartcontact of the (DC) full RCLsEPA RSL worksheets. webpage, Tables and that 1 andthe other 2 of thissubparts document that will summarize have important the DC-RCL explanatory changes text, generic tablesfrom and the references previous have spreadsheet yet to be (Januaryupdated. -

US EPA, Pesticide Product Label, Metolachlor + Metribuzin EC,04/15
U.S. ENVIRONMENTAL PROTECTION AGENCY EPA Reg. Number: Date of Issuance: Office of Pesticide Programs Registration Division (7505P) 42750-360 4/15/20 1200 Pennsylvania Ave., N.W. Washington, D.C. 20460 NOTICE OF PESTICIDE: Term of Issuance: X Registration Reregistration Conditional (under FIFRA, as amended) Name of Pesticide Product: METOLACHLOR + METRIBUZIN EC Name and Address of Registrant (include ZIP Code): Albaugh, LLC P.O. Box 2127 Valdosta, GA 31604-2127 Note: Changes in labeling differing in substance from that accepted in connection with this registration must be submitted to and accepted by the Registration Division prior to use of the label in commerce. In any correspondence on this product always refer to the above EPA registration number. On the basis of information furnished by the registrant, the above named pesticide is hereby registered under the Federal Insecticide, Fungicide and Rodenticide Act. Registration is in no way to be construed as an endorsement or recommendation of this product by the Agency. In order to protect health and the environment, the Administrator, on his motion, may at any time suspend or cancel the registration of a pesticide in accordance with the Act. The acceptance of any name in connection with the registration of a product under this Act is not to be construed as giving the registrant a right to exclusive use of the name or to its use if it has been covered by others. This product is conditionally registered in accordance with FIFRA section 3(c)(7)(A). You must comply with the following conditions: 1. Submit and/or cite all data required for registration/reregistration/registration review of your product under FIFRA when the Agency requires all registrants of similar products to submit such data. -

Nozzle Selection and Adjuvant Impact on the Efficacy of Glyphosate And
agronomy Article Nozzle Selection and Adjuvant Impact on the Efficacy of Glyphosate and PPO-Inhibiting Herbicide Tank-Mixtures Jesaelen G. Moraes 1,* , Thomas R. Butts 1 , Vitor M. Anunciato 2 , Joe D. Luck 3 , Wesley C. Hoffmann 4, Ulisses R. Antuniassi 5 and Greg R. Kruger 1 1 Department of Agronomy and Horticulture, University of Nebraska-Lincoln, North Platte, NE 69101, USA; [email protected] (T.R.B.); [email protected] (G.R.K.) 2 Department of Plant Protection, Sao Paulo State University, Botucatu, SP 18618687, Brazil; [email protected] 3 Department of Biological System Engineering, University of Nebraska-Lincoln, North Platte, NE 69101, USA; [email protected] 4 USDA-ARS Aerial Application Technology Research Unit, College Station, TX 77845, USA; [email protected] 5 Department of Rural Engineering, Sao Paulo State University, Botucatu, SP 18618687, Brazil; [email protected] * Correspondence: [email protected]; Tel.: +1-402-219-1674 Abstract: PPO-inhibiting herbicides in combination with glyphosate for postemergence applications is a common approach to manage glyphosate- and ALS-inhibitor-resistant weeds. PPO-inhibitors can reduce glyphosate translocation when applied in tank-mixtures, but adjuvants may be used to overcome this effect. Additionally, optimal droplet size may be affected by tank-mixtures of different Citation: Moraes, J.G.; Butts, T.R.; herbicides and it can be crucial to herbicide efficacy. Field and greenhouse studies were conducted M. Anunciato, V.; Luck, J.D.; to investigate the impact of nozzle selection and adjuvants on weed control and interactions when Hoffmann, W.C.; Antuniassi, U.R.; applying PPO-inhibitors (fomesafen or lactofen) alone or in tank-mixture with glyphosate to five Kruger, G.R. -
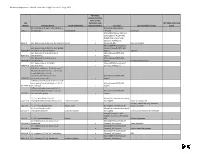
Chemicals of High Concern List (Sorted Alphabetically), July 2010
Minnesota Department of Health, Chemicals of High Concern list, July 1, 2010 Persistent, Bioaccumulative, Toxic or very CAS Persistent, very HPV (2006 and 3 of 4 Number Chemical Name Health endpoint(s) Bioaccumulative Source(s) Use example(s) or class years) (S)-4-hydroxy-3-(3-oxo-1-phenylbutyl)-2- Maine (EU Reproductive 5543-57-7 benzopyrone Reproduction Toxicant) Sunscreen Maine (CA Prop 65; IARC; EU Carcinogen; NTP 11th ROC; OSPAR Chemicals of High Concern); WA Appen1; 91-94-1 [1,1'-biphenyl]-4,4'-diamine, 3,3'-dichloro-Cancer x Minnesota HRL Dye, curing agent Maine (OSPAR Chemicals of [1,1'-biphenyl]-4,4'-diamine, N,N'-bis(2,4- Concern; Canada PBiT); WA 29398-96-7 dinitrophenyl)-3,3'-dimethoxy- x Appen1 Colorant [1,1'-Biphenyl]-4-ol, 3,4',5-tris(1,1- Maine (Canada PBiT); WA 6257-39-2 dimethylethyl)- x Appen1 [1,1'-Biphenyl]-4-ol, 3,4'-bis(1,1- Maine (Canada PBiT); WA 42479-88-9 dimethylethyl)- x Appen1 Chemical intermediate [1,1'-biphenyl]-4-ol, 3,5-bis(1,1- Maine (OSPAR Chemicals of 2668-47-5 dimethylethyl)- x Concern); WA Appen1 [2,6'-Bibenzothiazole]-7-sulfonic acid, 2'- (4-aminophenyl)-6-methyl-, diazotized, coupled with diazotized 4- aminobenzenesulfonic acid and Maine (Canada PBiT); WA 91696-90-1 resorcinol, sodium salts x Appen1 1(2H)-Quinolineethanol, 6-[(2-chloro-4,6- dinitrophenyl) azo]-3,4-dihydro-2,2,4,7- Maine (Canada PBiT); WA 63133-84-6 tetramethyl- x Appen1 1(2H)-Quinolinepropanamide, 6-(2,2- dicyanoethenyl)-3, 4-dihydro-2,2,4,7- Maine (Canada PBiT); WA 63467-15-2 tetramethyl-N-phenyl- x Appen1 1,1,1,2-Tetrachloro-2,2-bis(4- -

2019 Minnesota Chemicals of High Concern List
Minnesota Department of Health, Chemicals of High Concern List, 2019 Persistent, Bioaccumulative, Toxic (PBT) or very Persistent, very High Production CAS Bioaccumulative Use Example(s) and/or Volume (HPV) Number Chemical Name Health Endpoint(s) (vPvB) Source(s) Chemical Class Chemical1 Maine (CA Prop 65; IARC; IRIS; NTP Wood and textiles finishes, Cancer, Respiratory 11th ROC); WA Appen1; WA CHCC; disinfection, tissue 50-00-0 Formaldehyde x system, Eye irritant Minnesota HRV; Minnesota RAA preservative Gastrointestinal Minnesota HRL Contaminant 50-00-0 Formaldehyde (in water) system EU Category 1 Endocrine disruptor pesticide 50-29-3 DDT, technical, p,p'DDT Endocrine system Maine (CA Prop 65; IARC; IRIS; NTP PAH (chem-class) 11th ROC; OSPAR Chemicals of Concern; EuC Endocrine Disruptor Cancer, Endocrine Priority List; EPA Final PBT Rule for 50-32-8 Benzo(a)pyrene x x system TRI; EPA Priority PBT); Oregon P3 List; WA Appen1; Minnesota HRV WA Appen1; Minnesota HRL Dyes and diaminophenol mfg, wood preservation, 51-28-5 2,4-Dinitrophenol Eyes pesticide, pharmaceutical Maine (CA Prop 65; IARC; NTP 11th Preparation of amino resins, 51-79-6 Urethane (Ethyl carbamate) Cancer, Development ROC); WA Appen1 solubilizer, chemical intermediate Maine (CA Prop 65; IARC; IRIS; NTP Research; PAH (chem-class) 11th ROC; EPA Final PBT Rule for 53-70-3 Dibenzo(a,h)anthracene Cancer x TRI; WA PBT List; OSPAR Chemicals of Concern); WA Appen1; Oregon P3 List Maine (CA Prop 65; NTP 11th ROC); Research 53-96-3 2-Acetylaminofluorene Cancer WA Appen1 Maine (CA Prop 65; IARC; IRIS; NTP Lubricant, antioxidant, 55-18-5 N-Nitrosodiethylamine Cancer 11th ROC); WA Appen1 plastics stabilizer Maine (CA Prop 65; IRIS; NTP 11th Pesticide (EPA reg. -

New Herbicide Strategies for Weed Management in Pumpkin and Soybean and Potato Vine Desiccation
New Herbicide Strategies for Weed Management in Pumpkin and Soybean and Potato Vine Desiccation James Harrison Ferebee IV Thesis submitted to the faculty of the Virginia Polytechnic Institute and State University in partial fulfillment of the requirements for the degree of Master of Science in Life Sciences In Plant Pathology, Physiology, and Weed Science Charles W. Cahoon Co-Chair Michael L. Flessner Co-Chair David B. Langston November 29, 2018 Suffolk, VA Keywords: Crop tolerance, herbicide resistance management, Palmer amaranth i New Herbicide Strategies for Weed Management in Pumpkin and Soybean and Potato Vine Desiccation J. Harrison Ferebee IV Abstract Weed control and desiccation are routinely executed with herbicides. Potato vine desiccation facilitates harvest, improves skin set, and regulates tuber size. Saflufenacil, glufosinate, saflufenacil plus glufosinate, and carfentrazone plus glufosinate were compared to diquat applied at 43, 31, and 17% B potatoes; similar vine desiccation (14 days after treatment), skin set, and yield were noted amongst treatments. Residual herbicides are routinely used for weed control in pumpkin. Fluridone and acetochlor formulations applied preemergence were evaluated in direct-seeded pumpkin compared to other labeled herbicides. Fluridone resulted in total crop loss following heavy rainfall immediately after planting; less rainfall resulted in transient injury. Acetochlor formulations resulted in significant pumpkin injury (34 to 39%) 14 days after planting. S- metolachlor controlled weeds similar to acetochlor without significant injury. Palmer amaranth has developed resistance to six different herbicide modes of action. The weed grows rapidly and is best controlled <10 cm in height. To control glyphosate and ALS- resistant biotypes, fomesafen plus dicamba were applied at first postemergence (POST) to small Palmer amaranth (<5 cm, 0 d) and at simulated delays of 7, 14, 21, and 28 d. -

Chemical Weed Control
2014 North Carolina Agricultural Chemicals Manual The 2014 North Carolina Agricultural Chemicals Manual is published by the North Carolina Cooperative Extension Service, College of Agriculture and Life Sciences, N.C. State University, Raleigh, N.C. These recommendations apply only to North Carolina. They may not be appropriate for conditions in other states and may not comply with laws and regulations outside North Carolina. These recommendations are current as of November 2013. Individuals who use agricultural chemicals are responsible for ensuring that the intended use complies with current regulations and conforms to the product label. Be sure to obtain current information about usage regulations and examine a current product label before applying any chemical. For assistance, contact your county Cooperative Extension agent. The use of brand names and any mention or listing of commercial products or services in this document does not imply endorsement by the North Carolina Cooperative Extension Service nor discrimination against similar products or services not mentioned. VII — CHEMICAL WEED CONTROL 2014 North Carolina Agricultural Chemicals Manual VII — CHEMICAL WEED CONTROL Chemical Weed Control in Field Corn ...................................................................................................... 224 Weed Response to Preemergence Herbicides — Corn ........................................................................... 231 Weed Response to Postemergence Herbicides — Corn ........................................................................ -
Herbicide Classification Chart
HERBICIDE CLASSIFICATION Repeated use of herbicides with the same site of action can result in the development of herbicide-resistant weed populations. This chart groups herbicides by their modes of action to assist you in selecting This chart lists premix herbicides alphabetically by their trade names by MODE OF herbicides 1) to maintain greater diversity in herbicide use and 2) to rotate by PREMIX so you can identify the premix’s component herbicides and their respective ACTION among effective herbicides with different sites of action to delay the development site-of-action groups. Refer to the Site-of-Action chart on the left (effect on plant growth) of herbicide resistance. for more information. SITEOFACTION NUMBER OF RESISTANT COMPONENT COMPONENT GROUP WEED SPECIES IN U.S. SITEOFACTION SITEOFACTION GROUP GROUP CHEMICAL ACTIVE PRODUCT ACTIVE TRADE ACTIVE TRADE SITE OF ACTION FAMILY INGREDIENT EXAMPLES PREMIX ® PREMIX ® (TRADE NAME®) INGREDIENT NAME INGREDIENT NAME LIPID SYNTHESIS INHIBITORS bicyclopyrone ––––– 27 acetochlor Harness 15 HARNESS XTRA clodinafop Discover NG mesotrione Callisto 27 atrazine AAtrex 5 ACURON cyhalofop Clincher atrazine AAtrex 5 clopyralid Stinger 4 Aryloxyphenoxypropio- fenoxaprop Ricestar, Tecoma, others HORNET nate (fops) s-metolachlor Dual II Magnum 15 flumetsulam Python 2 ACCASE INHIBITORS fluazifop Fusilade DX bicyclopyrone ––––– 27 pyrasulfotole ––––– 27 1 (acetyl CoA carboxylase) 15 HUSKIE quizalofop Assure II, Targa Acuron Flexi mesotrione Callisto 27 bromoxynil Buctril 6 clethodim Select Max, others -

Droplet Size
Best Practices for Ground Application 1 Pesticide Spray Drift Series—3 Parts • March 15, 2018 webinar: “Strategies for Managing Pesticide Spray Drift” • Presented by Dr. Greg Kruger, University of Nebraska-Lincoln • Covers fundamentals of pesticide spray particle drift management • Materials available: https://www.epa.gov/reducing-pesticide-drift/strategies-managing- pesticide-spray-drift-webinar-materials • September 27, 2018 webinar: “Best Practices for Aerial Application” • Presented by Br. Bradley Fritz, United States Department of Agriculture • Dr. Greg Kruger joined for the Q+A discussion • Webinar materials will be posted online • Today’s webinar: “Best Practices for Ground Application” • Presented by Dr. Greg Kruger, University of Nebraska-Lincoln • Dr. Bradley Fritz will join for the Q+A discussion 4 Joining us for Q+A discussion • Bradley Fritz, Ph.D • Agricultural engineer and Research Leader, Agricultural Research Service, US Department of Agriculture • Research areas: examining the role of spray nozzles, spray solutions, and operational settings in resulting droplet size of spray; exploring the transport and fate of applied spray under field conditions • Numerous publications: https://www.ars.usda.gov/people- locations/person?person-id=33323 5 Presenter Greg Kruger, Ph.D. • Weed science and pesticide application technology specialist • University of Nebraska-Lincoln, Department of Agronomy and Horticulture • Director of the Pesticide Application Technology Laboratory • Areas of research: droplet size and efficacy, spray drift deposition and canopy penetration, influence of nozzle type, orifice size, spray pressure, and carrier volume rate on spray droplet size • Weed Science Society of America liaison to EPA 6 Best Practices for Ground Application Greg R. Kruger Weed Science and Application Technology Specialist WCREC, North Platte, NE NeOiasMa I I ' 8 Definition of Drift: Movement of spray particles and vapors off-target causing less effective control and possible injury to susceptible vegetation, wildlife, and people. -

400 Part 180—Tolerances And
Pt. 180 40 CFR Ch. I (7–1–11 Edition) (iv) The data and information sub- 180.4 Exceptions. mitted in support of the petition. 180.5 Zero tolerances. (v) The notice of filing of the peti- 180.6 Pesticide tolerances regarding milk, tion. eggs, meat, and/or poultry; statement of policy. (3) Any order issued under § 180.29(f) of this chapter to which the objection Subpart B—Procedural Regulations related, the regulation that was the subject of that order, and each related 180.7 Petitions proposing tolerances or ex- Notice of Proposed Rulemaking. emptions for pesticide residues in or on raw agricultural commodities or proc- (4) The comments submitted by mem- essed foods. bers of the public in response to the 180.8 Withdrawal of petitions without preju- Notice of Filing or Notice of Proposed dice. Rulemaking, and the information sub- 180.9 Substantive amendments to petitions. mitted as part of the comments, the 180.29 Establishment, modification, and rev- Administrator’s response to comments ocation of tolerance on initiative of Ad- and the documents or information re- ministrator. lied on by the Administrator in issuing 180.30 Judicial review. 180.31 Temporary tolerances. the regulation or order. 180.32 Procedure for modifying and revoking (5) All other documents or informa- tolerances or exemptions from toler- tion submitted to the docket for the ances. rulemaking in question under parts 177 180.33 Fees. or part 180 of this chapter. 180.34 Tests on the amount of residue re- (6) The Notice of Hearing published maining. under § 179.20.