Genetic Variations Associated with Six-White-Point Coat Pigmentation In
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Molecular Signature Induced by RNASET2, a Tumor Antagonizing Gene, in Ovarian Cancer Cells
www.impactjournals.com/oncotarget/ Oncotarget, June, Vol.2, No 6 Molecular signature induced by RNASET2, a tumor antagonizing gene, in ovarian cancer cells Francesco Acquati1, Laura Monti1, Marta Lualdi1, Marco Fabbri2, Maria Grazia Sacco2, Laura Gribaldo2, and Roberto Taramelli1 1 Dipartimento di Biotecnologie e Scienze Molecolari, Università degli Studi dell’Insubria, via JH Dunant 3, 21100 Varese, Italy 2 European Commission - Joint Research Centre Institute for Health and Consumer Protection Molecular Biology and Genomics unit TP 464, Via E. Fermi, 2749 21027 Ispra (VA) - Italy Correspondence to: Roberto Taramelli, email: [email protected] Correspondence to: Francesco Acquati, email: [email protected] Keywords: RNases, cancer microenvironment, transcriptional profile Received: May 10, 2011, Accepted: June 2, 2011, Published: June 4, 2011 Copyright: © Acquati et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT: Using the Hey3Met2 human ovarian cancer cell line, we previously found the RNASET2 gene to possess a remarkable in vivo tumor suppressor activity, although no in vitro features such as inhibition of cell proliferation, clonogenic potential, impaired growth in soft agar and increase in apoptotic rate could be detected. This is reminiscent of the behavior of genes belonging to the class of tumor antagonizing genes (TAG) which act mainly within the context of the microenvironment. Here we present transcriptional profiles analysis which indicates that investigations of the mechanisms of TAG biological functions require a comparison between the in vitro and in vivo expression patterns. -
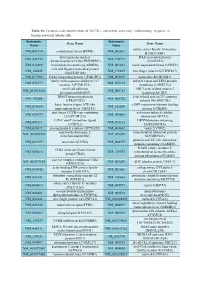
Systematic Name Gene Name Systematic Name Gene Name NM 001710 Complement Factor B(CFB) NM 052831 Solute Carrier Family 18 Member
Table S1: Genome-wide identification of SGLT2i`s interaction with early inflammatory response in human proximal tubular cells. Systematic Systematic Gene Name Gene Name Name Name solute carrier family 18 member NM_001710 complement factor B(CFB) NM_052831 B1(SLC18B1) heterogeneous nuclear DAZ associated protein NM_031372 NM_170711 ribonucleoprotein D like(HNRNPDL) 1(DAZAP1) NM_014299 bromodomain containing 4(BRD4) NM_001261 cyclin dependent kinase 9(CDK9) cilia and flagella associated protein NM_182628 NM_178835 zinc finger protein 827(ZNF827) 100(CFAP100) NM_017906 PAK1 interacting protein 1(PAK1IP1) NM_024015 homeobox B4(HOXB4) family with sequence similarity 167 ankyrin repeat and LEM domain NM_053279 NM_015114 member A(FAM167A) containing 2(ANKLE2) small cell adhesion ARP3 actin related protein 3 NM_001031628 NM_005721 glycoprotein(SMAGP) homolog(ACTR3) TRAF3 interacting protein actin related protein 2/3 complex NM_147686 NM_005720 2(TRAF3IP2) subunit 1B(ARPC1B) basic leucine zipper ATF-like cAMP responsive element binding NM_018664 NM_182898 transcription factor 3(BATF3) protein 5(CREB5) zinc finger CCCH-type containing activation induced cytidine NM_025079 NM_020661 12A(ZC3H12A) deaminase(AICDA) C-X-C motif chemokine ligand DENN domain containing NM_001511 NM_015213 1(CXCL1) 5A(DENND5A) NM_025072 prostaglandin E synthase 2(PTGES2) NM_004665 vanin 2(VNN2) superoxide dismutase 2, mitochondrial ribosomal protein NM_001024465 NM_016070 mitochondrial(SOD2) S23(MRPS23) jumonji and AT-rich interaction NM_033199 urocortin 2(UCN2) NM_004973 -

Therapeutic Vaccines for Amyotrophic Lateral Sclerosis Directed Against
Vaccine 37 (2019) 4920–4927 Contents lists available at ScienceDirect Vaccine journal homepage: www.elsevier.com/locate/vaccine Therapeutic vaccines for amyotrophic lateral sclerosis directed against disease specific epitopes of superoxide dismutase 1 ⇑ Beibei Zhao a, Kristen Marciniuk b,c, Ebrima Gibbs a, Masoud Yousefi a, Scott Napper b,c, Neil R. Cashman a, a Djavad Mowafaghian Centre for Brain Health, University of British Columbia, Vancouver, BC V6T 2B5, Canada b Department of Biochemistry, Microbiology, and Immunology, University of Saskatchewan, Saskatoon, Saskatchewan S7N 5E5, Canada c Vaccine and Infectious Disease Organization International Vaccine Research Center, University of Saskatchewan, Saskatchewan, SK S7N 5E3, Canada article info abstract Article history: Emerging evidence suggests seeding and prion-like propagation of mutant Superoxide Dismutase 1 Received 6 February 2019 (SOD1) misfolding to be a potential mechanism for ALS pathogenesis and progression. Immuno- Received in revised form 8 June 2019 targeting of misfolded SOD1 has shown positive clinical outcomes in mutant SOD1 transgenic mice. Accepted 10 July 2019 However, a major challenge in developing active immunotherapies for proteinopathies such as ALS is Available online 16 July 2019 the design of immunogens enabling exclusive recognition of pathogenic species of a self-protein. Ideally, one would achieve a robust antibody response against the disease-misfolded protein while spar- ing the natively folded conformer to avoid inducing deleterious autoimmune complications, or inhibiting its normal function. Using a motor neuron disease mouse model expressing human SOD1-G37R, we herein report the immunogenicity and therapeutic efficacy of two ALS vaccines, tgG-DSE2lim and tgG- DSE5b, based on the notion that native SOD1 would undergo early unfolding in disease to present ‘‘dis- ease specific epitopes” (DSE). -

A Genome Wide Association Study of Fast Beta EEG in Families of European Ancestry
CORE Metadata, citation and similar papers at core.ac.uk Provided by IUPUIScholarWorks HHS Public Access Author manuscript Author ManuscriptAuthor Manuscript Author Int J Psychophysiol Manuscript Author . Author Manuscript Author manuscript; available in PMC 2018 May 01. Published in final edited form as: Int J Psychophysiol. 2017 May ; 115: 74–85. doi:10.1016/j.ijpsycho.2016.12.008. A Genome Wide Association Study of Fast Beta EEG in Families of European Ancestry Jacquelyn L. Meyers1,*, Jian Zhang1, Niklas Manz1,2, Madhavi Rangaswamy3, Chella Kamarajan1, Leah Wetherill4, David B. Chorlian1, Sun J. Kang5, Lance Bauer6, Victor Hesselbrock6, John Kramer7, Samuel Kuperman7, John I. Nurnberger Jr4, Jay Tischfield8, Jen Chyong Wang9, Howard J. Edenberg4,10, Alison Goate9, Tatiana Foroud4, and Bernice Porjesz1 1Henri Begleiter Neurodynamics Laboratory, Department of Psychiatry and Behavioral Sciences, SUNY Downstate Medical Center, Brooklyn, NY, USA 2Department of Physics, College of Wooster, Wooster, OH, USA 3Department of Psychology, Christ University, Bangalore, India 4Department of Medical and Molecular Genetics, Indiana University School of Medicine, Indianapolis, IN, USA 5Albany Stratton VA Medical Center, Albany, NY, USA 6University of Connecticut School of Medicine, Farmington, CT, USA 7Department of Psychiatry, University of Iowa, Iowa City, IA, USA 8Rutgers University, Piscataway, NJ, USA 9Icahn School of Medicine at Mt. Sinai, New York, NY, USA 10Department of Biochemistry and Molecular Biology, Indiana University School of Medicine, Indianapolis, IN, USA Abstract BACKGROUND—Differences in fast beta (20–28 Hz) electroencephalogram (EEG) oscillatory activity distinguish some individuals with psychiatric and substance use disorders, suggesting that it may be a useful endophenotype for studying the genetics of disorders characterized by neural hyper-excitability. -

RNA: Transcription, Processing, and Decay 8
CHAPTER RNA: Transcription, Processing, and Decay 8 Knowledge of the molecular mechanisms that synthesize and destroy RNAs in cells has led to technologies that allow researchers to control gene expression in precise ways. For example, on the left is CHAPTER OUTLINE AND LEARNING OBJECTIVES a C. elegans worm that has been manipulated to express a gene encoding the green fluorescent protein 8.1 RNA STRUCTURE (GFP) in specific cells, and on the right LO 8.1 Describe how the structure of RNA enables it to function differently from DNA. is a genetically identical worm in which GFP expression is silenced. [ Jessica Vasale/Laboratory of Craig Mello. ] 8.2 TRANSCRIPTION AND DECAY OF mRNA IN BACTERIA LO 8.2 Explain how RNA polymerases are directed to begin and end transcription at specific places in genomes. 8.3 TRANSCRIPTION IN EUKARYOTES LO 8.3 Describe how mRNA transcription and decay mechanisms in eukaryotes are similar to those in bacteria. 8.4 PROCESSING OF mRNA IN EUKARYOTES LO 8.4 Explain how mRNA processing, editing, and modification occur and can affect the abundance and sequence of proteins in eukaryotes. 8.5 DECAY OF mRNA IN EUKARYOTES LO 8.5 Describe how siRNAs regulate the abundance of specific RNAs and play a role in maintaining genome integrity in eukaryotes. 267 11_GriffitITGA12e_11478_Ch08_267_300.indd 267 14/10/19 9:49 AM The broad objective for this chapter is to understand the mechanisms of RNA CHAPTER OBJECTIVE synthesis, processing, and decay as well as how RNA and protein factors reg- ulate these mechanisms. n this chapter, we describe how the information stored in eliminated from cells by decay mechanisms that disassem- DNA is transferred to RNA. -

Downloaded Per Proteome Cohort Via the Web- Site Links of Table 1, Also Providing Information on the Deposited Spectral Datasets
www.nature.com/scientificreports OPEN Assessment of a complete and classifed platelet proteome from genome‑wide transcripts of human platelets and megakaryocytes covering platelet functions Jingnan Huang1,2*, Frauke Swieringa1,2,9, Fiorella A. Solari2,9, Isabella Provenzale1, Luigi Grassi3, Ilaria De Simone1, Constance C. F. M. J. Baaten1,4, Rachel Cavill5, Albert Sickmann2,6,7,9, Mattia Frontini3,8,9 & Johan W. M. Heemskerk1,9* Novel platelet and megakaryocyte transcriptome analysis allows prediction of the full or theoretical proteome of a representative human platelet. Here, we integrated the established platelet proteomes from six cohorts of healthy subjects, encompassing 5.2 k proteins, with two novel genome‑wide transcriptomes (57.8 k mRNAs). For 14.8 k protein‑coding transcripts, we assigned the proteins to 21 UniProt‑based classes, based on their preferential intracellular localization and presumed function. This classifed transcriptome‑proteome profle of platelets revealed: (i) Absence of 37.2 k genome‑ wide transcripts. (ii) High quantitative similarity of platelet and megakaryocyte transcriptomes (R = 0.75) for 14.8 k protein‑coding genes, but not for 3.8 k RNA genes or 1.9 k pseudogenes (R = 0.43–0.54), suggesting redistribution of mRNAs upon platelet shedding from megakaryocytes. (iii) Copy numbers of 3.5 k proteins that were restricted in size by the corresponding transcript levels (iv) Near complete coverage of identifed proteins in the relevant transcriptome (log2fpkm > 0.20) except for plasma‑derived secretory proteins, pointing to adhesion and uptake of such proteins. (v) Underrepresentation in the identifed proteome of nuclear‑related, membrane and signaling proteins, as well proteins with low‑level transcripts. -

Lifestyle Intervention in Pregnant Women with Obesity Impacts Cord Blood DNA Methylation, Which Associates with Body Composition in the Offspring
854 Diabetes Volume 70, April 2021 Lifestyle Intervention in Pregnant Women With Obesity Impacts Cord Blood DNA Methylation, Which Associates With Body Composition in the Offspring Josefine Jönsson,1 Kristina M. Renault,2,3 Sonia García-Calzón,1,4 Alexander Perfilyev,1 Angela C. Estampador,5 Kirsten Nørgaard,6 Mads Vendelbo Lind,7 Allan Vaag,6 Line Hjort,8 Kim F. Michaelsen,7 Emma Malchau Carlsen,7,9 Paul W. Franks,5 and Charlotte Ling1 Diabetes 2021;70:854–866 | https://doi.org/10.2337/db20-0487 Maternal obesity may lead to epigenetic alterations in the mediates the effect of the lifestyle intervention on lean offspring and might thereby contribute to disease later in mass in the offspring (FDR <5%). Moreover, 22 methyla- life. We investigated whether a lifestyle intervention in tion sites were associated with offspring BMI z scores pregnant women with obesity is associated with epige- during the first 3 years of life (P < 0.05). Overall, lifestyle netic variation in cord blood and body composition in the interventions in pregnant women with obesity are asso- offspring. Genome-wide DNA methylation was analyzed ciated with epigenetic changes in offspring, potentially in cord blood from 208 offspring from the Treatment of influencing the offspring’s lean mass and early growth. Obese Pregnant women (TOP)-study, which includes pregnant women with obesity randomized to lifestyle interventions comprised of physical activity with or with- Obesity and type 2 diabetes are on the rise worldwide, as is out dietary advice versus control subjects (standard of the prevalence of obesity in pregnant women (1). -

Anthropology Under Graduate Curriculum Under Choice Based Credit System 2015 Overview of Curriculum
ANTHROPOLOGY UNDER GRADUATE CURRICULUM UNDER CHOICE BASED CREDIT SYSTEM 2015 OVERVIEW OF CURRICULUM I. Core course Year Semester Paper No. Title of Paper I ANTH-101 Introduction to Biological Anthropology First ANTH-102 Introduction to Socio-cultural Anthropology II ANTH-201 Archaeological Anthropology ANTH-202 Fundamentals of Human Origin & Evolution III ANTH-301 Tribes and Peasants in India ANTH-302 Human Ecology: Biological & Cultural dimensions Second ANTH-303 Biological Diversity in Human Populations IV ANTH-401 Theories of Culture and Society ANTH-402 Human Growth and Development ANTH-403 Research Methods V ANTH-501 Human Population Genetics Third ANTH-502 Anthropology in Practice VI ANTH-601 Forensic Anthropology ANTH-602 Anthropology of India II. Elective Course A. Discipline Specific B. Generic Elective/Interdisciplinary Two each in Semester V and VI. To be chosen from the One each in Semester I, II, III and IV. To be following. chosen from the following. DSE-1: Physiological Anthropology GE-1: Health science DSE-2: Sports and Nutritional Anthropology GE-2: Home science DSE-3: Human Genetics GE-3: Biotechnology DSE-4: Neuro Anthropology GE-4: Psychology DSE-5: Forensic Dermatoglyphics GE-5: Animation and Visual Graphics DSE-6: Paleoanthropology GE-6: Interior Design DSE-7: Anthropology of Religion, Politics and economy GE-7: Economics DSE-8: Tribal Cultures of India GE-8: Environmental Science DSE-9: Indian Archaeology GE-9: Fashion Design DSE-10: Visual Anthropology GE-10: Food Technology DSE-11: Fashion Anthropology GE-11: Forestry DSE-12: Demographic Anthropology GE-12: Neuro Science DSE-13: Urban Anthropology GE-13: Physical Education DSE-14: Anthropology of Health GE-14: Tourism Administration DSE-15: Dissertation (in Semester VI only) GE-15: Insurance and Banking GE-16: Journalism and Mass Communication GE-17: BCA GE-18: BBA GE-19: Hotel Management GE-20: BBA (Health Care Management) GE-21: Marine Science III. -

Autocrine IFN Signaling Inducing Profibrotic Fibroblast Responses By
Downloaded from http://www.jimmunol.org/ by guest on September 23, 2021 Inducing is online at: average * The Journal of Immunology , 11 of which you can access for free at: 2013; 191:2956-2966; Prepublished online 16 from submission to initial decision 4 weeks from acceptance to publication August 2013; doi: 10.4049/jimmunol.1300376 http://www.jimmunol.org/content/191/6/2956 A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Autocrine IFN Signaling Feng Fang, Kohtaro Ooka, Xiaoyong Sun, Ruchi Shah, Swati Bhattacharyya, Jun Wei and John Varga J Immunol cites 49 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2013/08/20/jimmunol.130037 6.DC1 This article http://www.jimmunol.org/content/191/6/2956.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 23, 2021. The Journal of Immunology A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Inducing Autocrine IFN Signaling Feng Fang,* Kohtaro Ooka,* Xiaoyong Sun,† Ruchi Shah,* Swati Bhattacharyya,* Jun Wei,* and John Varga* Activation of TLR3 by exogenous microbial ligands or endogenous injury-associated ligands leads to production of type I IFN. -

Supplemental Figures 04 12 2017
Jung et al. 1 SUPPLEMENTAL FIGURES 2 3 Supplemental Figure 1. Clinical relevance of natural product methyltransferases (NPMTs) in brain disorders. (A) 4 Table summarizing characteristics of 11 NPMTs using data derived from the TCGA GBM and Rembrandt datasets for 5 relative expression levels and survival. In addition, published studies of the 11 NPMTs are summarized. (B) The 1 Jung et al. 6 expression levels of 10 NPMTs in glioblastoma versus non‐tumor brain are displayed in a heatmap, ranked by 7 significance and expression levels. *, p<0.05; **, p<0.01; ***, p<0.001. 8 2 Jung et al. 9 10 Supplemental Figure 2. Anatomical distribution of methyltransferase and metabolic signatures within 11 glioblastomas. The Ivy GAP dataset was downloaded and interrogated by histological structure for NNMT, NAMPT, 12 DNMT mRNA expression and selected gene expression signatures. The results are displayed on a heatmap. The 13 sample size of each histological region as indicated on the figure. 14 3 Jung et al. 15 16 Supplemental Figure 3. Altered expression of nicotinamide and nicotinate metabolism‐related enzymes in 17 glioblastoma. (A) Heatmap (fold change of expression) of whole 25 enzymes in the KEGG nicotinate and 18 nicotinamide metabolism gene set were analyzed in indicated glioblastoma expression datasets with Oncomine. 4 Jung et al. 19 Color bar intensity indicates percentile of fold change in glioblastoma relative to normal brain. (B) Nicotinamide and 20 nicotinate and methionine salvage pathways are displayed with the relative expression levels in glioblastoma 21 specimens in the TCGA GBM dataset indicated. 22 5 Jung et al. 23 24 Supplementary Figure 4. -

Transcriptomics of Cumulus Cells – a Window Into Oocyte Maturation in Humans Brandon A
Wyse et al. Journal of Ovarian Research (2020) 13:93 https://doi.org/10.1186/s13048-020-00696-7 RESEARCH Open Access Transcriptomics of cumulus cells – a window into oocyte maturation in humans Brandon A. Wyse1*† , Noga Fuchs Weizman1†, Seth Kadish1, Hanna Balakier1, Mugundhine Sangaralingam1 and Clifford L. Librach1,2,3,4 Abstract Background: Cumulus cells (CC) encapsulate growing oocytes and support their growth and development. Transcriptomic signatures of CC have the potential to serve as valuable non-invasive biomarkers for oocyte competency and potential. The present sibling cumulus-oocyte-complex (COC) cohort study aimed at defining functional variations between oocytes of different maturity exposed to the same stimulation conditions, by assessing the transcriptomic signatures of their corresponding CC. CC were collected from 18 patients with both germinal vesicle and metaphase II oocytes from the same cycle to keep the biological variability between samples to a minimum. RNA sequencing, differential expression, pathway analysis, and leading-edge were performed to highlight functional differences between CC encapsulating oocytes of different maturity. Results: Transcriptomic signatures representing CC encapsulating oocytes of different maturity clustered separately on principal component analysis with 1818 genes differentially expressed. CCs encapsulating mature oocytes were more transcriptionally synchronized when compared with CCs encapsulating immature oocytes. Moreover, the transcriptional activity was lower, albeit not absent, in CC encapsulating mature oocytes, with 2407 fewer transcripts detected than in CC encapsulating immature (germinal vesicle - GV) oocytes. Hallmark pathways and ovarian processes that were affected by oocyte maturity included cell cycle regulation, steroid metabolism, apoptosis, extracellular matrix remodeling, and inflammation. Conclusions: Herein we review our findings and discuss how they align with previous literature addressing transcriptomic signatures of oocyte maturation. -

Identification of Epigenetic Mechanisms Involved in the Anti-Asthmatic Effects of Descurainia Sophia Seed Extract Based on a Multi-Omics Approach
molecules Article Identification of Epigenetic Mechanisms Involved in the Anti-Asthmatic Effects of Descurainia sophia Seed Extract Based on a Multi-Omics Approach Su-Jin Baek 1, Jin Mi Chun 2, Tae-Wook Kang 1, Yun-Soo Seo 2, Sung-Bae Kim 2, Boseok Seong 3, Yunji Jang 3, Ga-Hee Shin 1 and Chul Kim 3,* 1 Bioinformatics Group, R&D Center, Insilicogen Corporation, 35, Techno 9-ro, Yuseong-gu, Daejeon 34027, Korea; [email protected] (S.-J.B.); [email protected] (T.-W.K.); [email protected] (G.-H.S.) 2 Herbal Medicine Research Division, Korea Institute of Oriental Medicine, 1672 Yuseong-daero, Yuseong-gu, Daejeon 34054, Korea; [email protected] (J.M.C.); [email protected] (Y.-S.S.); [email protected] (S.-B.K.) 3 Future Medicine Division, Korea Institute of Oriental Medicine, 1672 Yuseong-daero, Yuseong-gu, Daejeon 34054, Korea; [email protected] (B.S.); [email protected] (Y.J.) * Correspondence: [email protected]; Tel.: +82-42-868-9582 Received: 15 October 2018; Accepted: 30 October 2018; Published: 5 November 2018 Abstract: Asthma, a heterogeneous disease of the airways, is common around the world, but little is known about the molecular mechanisms underlying the interactions between DNA methylation and gene expression in relation to this disease. The seeds of Descurainia sophia are traditionally used to treat coughs, asthma and edema, but their effects on asthma have not been investigated by multi-omics analysis. We undertook this study to assess the epigenetic effects of ethanol extract of D. sophia seeds (DSE) in an ovalbumin (OVA)-induced mouse model of asthma.