Proximal Sequence Element-Binding Transcription Factor
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

L2-Transcription.Pdf
Transcription BIOL201 Daniel Gautheret [email protected] • Chapitre 1: Transcription et RNA polymérase • Chapitre 2: Initiation, élongation et terminaison • Chapitre 3: Régulation: l’exemple procaryote • Chapitre 4: Maturation de l’ARN chez les eucaryotes V. 2012.0 Chapitre 1 Transcription et RNA polymérase 2 L’intuition de la transcription Chez les eucaryotes: ADN dans le noyau Machinerie de synthèse dans le cytoplasme Hypothèse d’un l’intermédiaire ARN 3 Etapes-clé de la découverte de l’ARN messager • 1957: concept d’un « adaptateur ARN » (Crick) • 1956-57: Découverte progressive d’une nouvelle fraction d’ARN, polymorphe • 1959-60: découverte d’une « ARN polymérase » capable de synthétiser de l’ARN à partir d’ADN. • 1961. Mise au point des techniques d’hybridation ADN- ARN sur filtre de nitrocellulose. -> l’ARN est complémentaire d’un seul brin d’ADN • 1961. Démonstration de l’existence d’un ARN messager (Jacob & Monod) • 1961. Purification d’ARN polymérase bactérienne et synthèse in vitro d’ARN à partir de matrice ADN 4 Rappel: différences ADN/ARN ARN ADN ribose désoxyribose 5 Bases de l’ADN et de l’ARN Base puriques ADN: T ARN: U Base pyrimidiques 6 Structure des ARN • Molécule ordonnée linéaire O • Orientée 5’P->3’OH P • Grand nombre de O H2C structures secondaires possibles 7 Décodage du gène dans la cellule bactérienne ADN ARN polymérase Ribosome grande sous-Unité ribosome ARNm Ribosome petite sous-Unité Protéine ARNr ARNt aminoacides Membrane cellulaire 8 Les principaux ARN dans une bactérie • ARN ribosomiques (ARNr) -

Structure and Evolution of Four POU Domain Genes Expressed in Mouse Brain (POU Bnain-/POU Brain-2/POU Brain4/POU Scip/Homeobox) YOSHINOBU HARA*, ALESSANDRA C
Proc. Natl. Acad. Sci. USA Vol. 89, pp. 3280-3284, April 1992 Neurobiology Structure and evolution of four POU domain genes expressed in mouse brain (POU Bnain-/POU Brain-2/POU Brain4/POU Scip/homeobox) YOSHINOBU HARA*, ALESSANDRA C. ROVESCALLI, YONGSOK KIM, AND MARSHALL NIRENBERG Laboratory of Biochemical Genetics, National Heart, Lung, and Blood Institute, National Institutes of Health, Bethesda, MD 20892 Contributed by Marshall Nirenberg, January 6, 1992 ABSTRACT Four mouse POU domain genomic DNA acid sequences of the proteins are related to one another and clones-Brain-i, Brain-2, Brain-4, and Scip-and Bran-2 suggests that the genes originated by duplication of an cDNA, which are expressed in adult brain, were cloned and the ancestral class III POU domain gene.t coding and noncoding regions ofthe genes were sequenced. The amino acid sequences of the four PoU domains are highly conserved; sequences in other regions of the proteins also are MATERIALS AND METHODS conserved but to a lesser extent. The absence of introns from DNA Coming and Sequencing. Our objective was to clone the coding regions of the four POU domain genes and the POU domain genes expressed in mouse brain. Mixtures of similarity of amino acid sequences of the corresponding pro- oligodeoxynucleotides that correspond to highly conserved teins suggest that the coding region of the ancestral class HI amino acid sequences in POU domains were used as primers POU domain gene lacked introns and therefore may have for amplification ofadult mouse brain POU domain cDNA by originated by reverse transcription of a molecule of POU PCR, and the amplified DNA fragments were cloned. -

BRN2 Is a Transcriptional Repressor of CDH13 (T-Cadherin)
Laboratory Investigation (2012) 92, 1788–1800 & 2012 USCAP, Inc All rights reserved 0023-6837/12 $32.00 BRN2 is a transcriptional repressor of CDH13 (T-cadherin) in melanoma cells Lisa Ellmann1, Manjunath B Joshi2,3, Therese J Resink2, Anja K Bosserhoff1 and Silke Kuphal1 T-cadherin (cadherin 13, H-cadherin, gene name CDH13) has been proposed to act as a tumor-suppressor gene as its expression is significantly diminished in several types of carcinomas, including melanomas. Allelic loss and promoter hypermethylation have been proposed as mechanisms for silencing of CDH13. However, they do not account for loss of T-cadherin expression in all carcinomas, and other genetic or epigenetic alterations can be presumed. The present study investigated transcriptional regulation of CDH13 in melanoma. Bioinformatical analysis pointed to the presence of known BRN2 (also known as POU3F2 and N-Oct-3)-binding motifs in the CDH13 promoter sequence. We found an inverse correlation between BRN2 and T-cadherin protein and transcript expression. Reporter gene analysis and electrophoretic mobility shift assays in melanoma cells demonstrated that CDH13 is a direct target of BRN2 and that BRN2 is a functional transcriptional repressor of CDH13 promoter activity. The regulatory binding element of BRN2 was located À 219 bp of the CDH13 promoter proximal to the start codon and was identified as 50-CATGCAAAA-30. Ectopic expression of BRN2 in BRN2-negative/T-cadherin-positive melanoma cells resulted in suppression of CDH13 promoter activity, whereas BRN2 knockdown in BRN2-positive/T-cadherin-negative melanoma cells resulted in re-expression of T-cadherin transcripts and protein. -

POU Domain Family Values: Flexibility, Partnerships, and Developmental Codes
Downloaded from genesdev.cshlp.org on October 1, 2021 - Published by Cold Spring Harbor Laboratory Press REVIEW POU domain family values: flexibility, partnerships, and developmental codes Aimee K. Ryan and Michael G. Rosenfeld 1 Howard Hughes Medical Institute, Department and School of Medicine, University of California at San Diego, La Jolla, Califiornia 92093-0648 USA Transcription factors serve critical roles in the progres- primary focus of this review. Crystallization of the Oct-1 sive development of general body plan, organ commit- and Pit-1 POU domains on DNA and in vitro studies ment, and finally, specific cell types. It has been the hope examining the specificity of POU domain cofactor inter- of most developmental biologists that comparison of the actions suggest that the flexibility with which the POU biological roles of a series of individual members within domain recognizes DNA-binding sites is a critical com- a family will permit at least some predictive generaliza- ponent of its ability to regulate gene expression. The tions regarding the developmental events that are likely implications of these data with respect to the mecha- to be regulated by a particular class of transcription fac- nisms utilized by the POU domain family of transcrip- tors. Here, we present an overview of the developmental tion factors to control developmental events will also be functions of the family of transcription factors charac- discussed. terized by the POU DNA-binding motif afforded by re- cent in vivo studies. Conformation of the POU domain on DNA The POU domain family of transcription factors was defined following the observation that the products of High-affinity site-specific DNA binding by POU domain three mammalian genes, Pit-l, Oct-l, and Oct-2 and the transcription factors requires both the POU-specific do- protein encoded by the Caenorhabditis elegans gene main and the POU homeodomain (Sturm and Herr 1988; unc-86 shared a region of homology, known as the POU Ingraham et al. -

Significance of Wilms' Tumor Gene 1 As a Biomarker in Acute Leukemia and Solid Tumors
UMEÅ UNIVERSITY MEDICAL DISSERTATIONS New Series No. 1799 ISSN 0346-6612 ISBN 978-91-7601-458-5 Significance of Wilms’ Tumor Gene 1 as a Biomarker in Acute Leukemia and Solid Tumors Charlotta Andersson Department of Medical Biosciences, Clinical Chemistry and Pathology Umeå University, Sweden Umeå 2016 Copyright © 2016 Charlotta Andersson ISBN: 978-91-7601-458-5 ISSN: 0346-6612 Printed by: Print & Media Umeå, Sweden, 2016 In memory of Aihong Li Table of Contents Table of Contents i Abstract iii Abbreviations iv List of Papers v Introduction 1 WT1 structure 1 WT1 target genes 3 WT1 protein partners 5 WT1 in normal and abnormal development 5 Hematopoiesis and WT1 7 WT1 as a tumor suppressor gene 7 WT1 as an oncogene or a chameleon gene 7 Acute leukemia (AL) and WT1 in AL 8 Renal cell carcinoma (RCC) and WT1 in RCC 13 Ovarian carcinoma (OC) and WT1 in OC 15 Aims of the Thesis 20 Materials and Methods 21 Patients and tissue samples (Papers I-IV) 21 Classification and risk group stratification in AL (Papers I and IV) 22 ccRCC grade and stage (Paper II) 22 OC classification, grade and stage (Paper III) 22 Genomic DNA preparation, RNA extraction and cDNA preparation (Papers I, II and IV) 23 Quantitative assessment of WT1 transcript expression with real-time quantitative PCR (RQ-PCR) (Papers I and II) 23 Sequencing analysis of the WT1 gene (Papers II and IV) 24 Immunohistochemistry (IHC) (Paper III) 24 Enzyme-linked immunosorbent assay (ELISA) (Paper III) 25 Statistical analysis 25 Results and Discussion 27 Paper I 27 Paper II 31 Paper III 35 Paper IV 38 Conclusions 42 Acknowledgments 44 References 47 i ii Abstract Wilms’ tumor gene 1 (WT1) is a zinc finger transcriptional regulator with crucial functions in embryonic development. -

Molecular Signature Induced by RNASET2, a Tumor Antagonizing Gene, in Ovarian Cancer Cells
www.impactjournals.com/oncotarget/ Oncotarget, June, Vol.2, No 6 Molecular signature induced by RNASET2, a tumor antagonizing gene, in ovarian cancer cells Francesco Acquati1, Laura Monti1, Marta Lualdi1, Marco Fabbri2, Maria Grazia Sacco2, Laura Gribaldo2, and Roberto Taramelli1 1 Dipartimento di Biotecnologie e Scienze Molecolari, Università degli Studi dell’Insubria, via JH Dunant 3, 21100 Varese, Italy 2 European Commission - Joint Research Centre Institute for Health and Consumer Protection Molecular Biology and Genomics unit TP 464, Via E. Fermi, 2749 21027 Ispra (VA) - Italy Correspondence to: Roberto Taramelli, email: [email protected] Correspondence to: Francesco Acquati, email: [email protected] Keywords: RNases, cancer microenvironment, transcriptional profile Received: May 10, 2011, Accepted: June 2, 2011, Published: June 4, 2011 Copyright: © Acquati et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT: Using the Hey3Met2 human ovarian cancer cell line, we previously found the RNASET2 gene to possess a remarkable in vivo tumor suppressor activity, although no in vitro features such as inhibition of cell proliferation, clonogenic potential, impaired growth in soft agar and increase in apoptotic rate could be detected. This is reminiscent of the behavior of genes belonging to the class of tumor antagonizing genes (TAG) which act mainly within the context of the microenvironment. Here we present transcriptional profiles analysis which indicates that investigations of the mechanisms of TAG biological functions require a comparison between the in vitro and in vivo expression patterns. -

Wilms' Tumor Gene 1 in Different Types of Cancer
UMEÅ UNIVERSITY MEDICAL DISSERTATIONS New Series No.1717 ISSN 0346-6612 ISBN 978-91-7601-263-5 Wilms’ tumor gene 1 in different types of cancer Xingru Li Department of Medical Biosciences, Clinical Chemistry Umeå University, Sweden Umeå 2015 Copyright © 2015 Xingru Li ISBN: 978-91-7601-263-5 ISSN: 0346-6612 Printed by: Print & Media Umeå, Sweden, 2015 To my family Table of Contents Abstract .......................................................................................................... 1 Original Articles ............................................................................................. 2 Abbreviations ................................................................................................. 3 Introduction .................................................................................................... 4 WT1 (Wilms’ tumor gene 1) ...................................................................... 4 Structure of WT1 ..................................................................................... 4 WT1, the transcription factor ................................................................. 5 WT1 and its interacting partners ............................................................ 5 WT1 function .......................................................................................... 8 The tumor suppressor ........................................................................ 8 An oncogene ...................................................................................... 8 Mutations and -

Molecular Biology, Third Edition Neuroscience, Second Edition Plant Biology, Second Edition Sport & Exercise Biomechanics Sport & Exercise Physiology
Molecular Biology Third Edition ii Section K – Lipid metabolism BIOS INSTANT NOTES Series Editor: B.D. Hames, School of Biochemistry and Microbiology, University of Leeds, Leeds, UK Biology Animal Biology, Second Edition Biochemistry, Third Edition Bioinformatics Chemistry for Biologists, Second Edition Developmental Biology Ecology, Second Edition Genetics, Second Edition Immunology, Second Edition Mathematics & Statistics for Life Scientists Medical Microbiology Microbiology, Second Edition Molecular Biology, Third Edition Neuroscience, Second Edition Plant Biology, Second Edition Sport & Exercise Biomechanics Sport & Exercise Physiology Chemistry Consulting Editor: Howard Stanbury Analytical Chemistry Inorganic Chemistry, Second Edition Medicinal Chemistry Organic Chemistry, Second Edition Physical Chemistry Psychology Sub-series Editor: Hugh Wagner, Dept of Psychology, University of Central Lancashire, Preston, UK Cognitive Psychology Physiological Psychology Psychology Sport & Exercise Psychology Molecular Biology Third Edition Phil Turner, Alexander McLennan, Andy Bates & Mike White School of Biological Sciences, University of Liverpool, Liverpool, UK Published by: Taylor & Francis Group In US: 270 Madison Avenue New York, NY 10016 In UK: 4 Park Square, Milton Park Abingdon, OX14 4RN © 2005 by Taylor & Francis Group This edition published in the Taylor & Francis e-Library, 2007. “To purchase your own copy of this or any of Taylor & Francis or Routledge’s collection of thousands of eBooks please go to www.eBookstore.tandf.co.uk.” First edition published in 1997 Second edition published in 2000 Third edition published in 2005 ISBN 0–203–96732–1 Master e-book ISBN ISBN: 0-415-35167-7 (Print Edition) This book contains information obtained from authentic and highly regarded sources. Reprinted material is quoted with permission, and sources are indicated. -

The Oct1 Homolog Nubbin Is a Repressor of NF-Κb-Dependent Immune Gene Expression That Increases the Tolerance to Gut Microbiota
Dantoft et al. BMC Biology 2013, 11:99 http://www.biomedcentral.com/1741-7007/11/99 RESEARCH ARTICLE Open Access The Oct1 homolog Nubbin is a repressor of NF-κB-dependent immune gene expression that increases the tolerance to gut microbiota Widad Dantoft1, Monica M Davis1, Jessica M Lindvall2, Xiongzhuo Tang1, Hanna Uvell1,3, Anna Junell1, Anne Beskow1,4 and Ylva Engström1* Abstract Background: Innate immune responses are evolutionarily conserved processes that provide crucial protection against invading organisms. Gene activation by potent NF-κB transcription factors is essential both in mammals and Drosophila during infection and stress challenges. If not strictly controlled, this potent defense system can activate autoimmune and inflammatory stress reactions, with deleterious consequences for the organism. Negative regulation to prevent gene activation in healthy organisms, in the presence of the commensal gut flora, is however not well understood. Results: We show that the Drosophila homolog of mammalian Oct1/POU2F1 transcription factor, called Nubbin (Nub), is a repressor of NF-κB/Relish-driven antimicrobial peptide gene expression in flies. In nub1 mutants, which lack Nub-PD protein, excessive expression of antimicrobial peptide genes occurs in the absence of infection, leading to a significant reduction of the numbers of cultivatable gut commensal bacteria. This aberrant immune gene expression was effectively blocked by expression of Nub from a transgene. We have identified an upstream regulatory region, containing a cluster of octamer sites, which is required for repression of antimicrobial peptide gene expression in healthy flies. Chromatin immunoprecipitation experiments demonstrated that Nub binds to octamer-containing promoter fragments of several immune genes. -
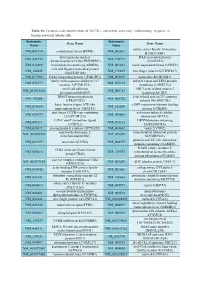
Systematic Name Gene Name Systematic Name Gene Name NM 001710 Complement Factor B(CFB) NM 052831 Solute Carrier Family 18 Member
Table S1: Genome-wide identification of SGLT2i`s interaction with early inflammatory response in human proximal tubular cells. Systematic Systematic Gene Name Gene Name Name Name solute carrier family 18 member NM_001710 complement factor B(CFB) NM_052831 B1(SLC18B1) heterogeneous nuclear DAZ associated protein NM_031372 NM_170711 ribonucleoprotein D like(HNRNPDL) 1(DAZAP1) NM_014299 bromodomain containing 4(BRD4) NM_001261 cyclin dependent kinase 9(CDK9) cilia and flagella associated protein NM_182628 NM_178835 zinc finger protein 827(ZNF827) 100(CFAP100) NM_017906 PAK1 interacting protein 1(PAK1IP1) NM_024015 homeobox B4(HOXB4) family with sequence similarity 167 ankyrin repeat and LEM domain NM_053279 NM_015114 member A(FAM167A) containing 2(ANKLE2) small cell adhesion ARP3 actin related protein 3 NM_001031628 NM_005721 glycoprotein(SMAGP) homolog(ACTR3) TRAF3 interacting protein actin related protein 2/3 complex NM_147686 NM_005720 2(TRAF3IP2) subunit 1B(ARPC1B) basic leucine zipper ATF-like cAMP responsive element binding NM_018664 NM_182898 transcription factor 3(BATF3) protein 5(CREB5) zinc finger CCCH-type containing activation induced cytidine NM_025079 NM_020661 12A(ZC3H12A) deaminase(AICDA) C-X-C motif chemokine ligand DENN domain containing NM_001511 NM_015213 1(CXCL1) 5A(DENND5A) NM_025072 prostaglandin E synthase 2(PTGES2) NM_004665 vanin 2(VNN2) superoxide dismutase 2, mitochondrial ribosomal protein NM_001024465 NM_016070 mitochondrial(SOD2) S23(MRPS23) jumonji and AT-rich interaction NM_033199 urocortin 2(UCN2) NM_004973 -

Therapeutic Vaccines for Amyotrophic Lateral Sclerosis Directed Against
Vaccine 37 (2019) 4920–4927 Contents lists available at ScienceDirect Vaccine journal homepage: www.elsevier.com/locate/vaccine Therapeutic vaccines for amyotrophic lateral sclerosis directed against disease specific epitopes of superoxide dismutase 1 ⇑ Beibei Zhao a, Kristen Marciniuk b,c, Ebrima Gibbs a, Masoud Yousefi a, Scott Napper b,c, Neil R. Cashman a, a Djavad Mowafaghian Centre for Brain Health, University of British Columbia, Vancouver, BC V6T 2B5, Canada b Department of Biochemistry, Microbiology, and Immunology, University of Saskatchewan, Saskatoon, Saskatchewan S7N 5E5, Canada c Vaccine and Infectious Disease Organization International Vaccine Research Center, University of Saskatchewan, Saskatchewan, SK S7N 5E3, Canada article info abstract Article history: Emerging evidence suggests seeding and prion-like propagation of mutant Superoxide Dismutase 1 Received 6 February 2019 (SOD1) misfolding to be a potential mechanism for ALS pathogenesis and progression. Immuno- Received in revised form 8 June 2019 targeting of misfolded SOD1 has shown positive clinical outcomes in mutant SOD1 transgenic mice. Accepted 10 July 2019 However, a major challenge in developing active immunotherapies for proteinopathies such as ALS is Available online 16 July 2019 the design of immunogens enabling exclusive recognition of pathogenic species of a self-protein. Ideally, one would achieve a robust antibody response against the disease-misfolded protein while spar- ing the natively folded conformer to avoid inducing deleterious autoimmune complications, or inhibiting its normal function. Using a motor neuron disease mouse model expressing human SOD1-G37R, we herein report the immunogenicity and therapeutic efficacy of two ALS vaccines, tgG-DSE2lim and tgG- DSE5b, based on the notion that native SOD1 would undergo early unfolding in disease to present ‘‘dis- ease specific epitopes” (DSE). -

A Genome Wide Association Study of Fast Beta EEG in Families of European Ancestry
CORE Metadata, citation and similar papers at core.ac.uk Provided by IUPUIScholarWorks HHS Public Access Author manuscript Author ManuscriptAuthor Manuscript Author Int J Psychophysiol Manuscript Author . Author Manuscript Author manuscript; available in PMC 2018 May 01. Published in final edited form as: Int J Psychophysiol. 2017 May ; 115: 74–85. doi:10.1016/j.ijpsycho.2016.12.008. A Genome Wide Association Study of Fast Beta EEG in Families of European Ancestry Jacquelyn L. Meyers1,*, Jian Zhang1, Niklas Manz1,2, Madhavi Rangaswamy3, Chella Kamarajan1, Leah Wetherill4, David B. Chorlian1, Sun J. Kang5, Lance Bauer6, Victor Hesselbrock6, John Kramer7, Samuel Kuperman7, John I. Nurnberger Jr4, Jay Tischfield8, Jen Chyong Wang9, Howard J. Edenberg4,10, Alison Goate9, Tatiana Foroud4, and Bernice Porjesz1 1Henri Begleiter Neurodynamics Laboratory, Department of Psychiatry and Behavioral Sciences, SUNY Downstate Medical Center, Brooklyn, NY, USA 2Department of Physics, College of Wooster, Wooster, OH, USA 3Department of Psychology, Christ University, Bangalore, India 4Department of Medical and Molecular Genetics, Indiana University School of Medicine, Indianapolis, IN, USA 5Albany Stratton VA Medical Center, Albany, NY, USA 6University of Connecticut School of Medicine, Farmington, CT, USA 7Department of Psychiatry, University of Iowa, Iowa City, IA, USA 8Rutgers University, Piscataway, NJ, USA 9Icahn School of Medicine at Mt. Sinai, New York, NY, USA 10Department of Biochemistry and Molecular Biology, Indiana University School of Medicine, Indianapolis, IN, USA Abstract BACKGROUND—Differences in fast beta (20–28 Hz) electroencephalogram (EEG) oscillatory activity distinguish some individuals with psychiatric and substance use disorders, suggesting that it may be a useful endophenotype for studying the genetics of disorders characterized by neural hyper-excitability.