RNA: Transcription, Processing, and Decay 8
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Proteomics Provides Insights Into the Inhibition of Chinese Hamster V79
www.nature.com/scientificreports OPEN Proteomics provides insights into the inhibition of Chinese hamster V79 cell proliferation in the deep underground environment Jifeng Liu1,2, Tengfei Ma1,2, Mingzhong Gao3, Yilin Liu4, Jun Liu1, Shichao Wang2, Yike Xie2, Ling Wang2, Juan Cheng2, Shixi Liu1*, Jian Zou1,2*, Jiang Wu2, Weimin Li2 & Heping Xie2,3,5 As resources in the shallow depths of the earth exhausted, people will spend extended periods of time in the deep underground space. However, little is known about the deep underground environment afecting the health of organisms. Hence, we established both deep underground laboratory (DUGL) and above ground laboratory (AGL) to investigate the efect of environmental factors on organisms. Six environmental parameters were monitored in the DUGL and AGL. Growth curves were recorded and tandem mass tag (TMT) proteomics analysis were performed to explore the proliferative ability and diferentially abundant proteins (DAPs) in V79 cells (a cell line widely used in biological study in DUGLs) cultured in the DUGL and AGL. Parallel Reaction Monitoring was conducted to verify the TMT results. γ ray dose rate showed the most detectable diference between the two laboratories, whereby γ ray dose rate was signifcantly lower in the DUGL compared to the AGL. V79 cell proliferation was slower in the DUGL. Quantitative proteomics detected 980 DAPs (absolute fold change ≥ 1.2, p < 0.05) between V79 cells cultured in the DUGL and AGL. Of these, 576 proteins were up-regulated and 404 proteins were down-regulated in V79 cells cultured in the DUGL. KEGG pathway analysis revealed that seven pathways (e.g. -

Molecular Signature Induced by RNASET2, a Tumor Antagonizing Gene, in Ovarian Cancer Cells
www.impactjournals.com/oncotarget/ Oncotarget, June, Vol.2, No 6 Molecular signature induced by RNASET2, a tumor antagonizing gene, in ovarian cancer cells Francesco Acquati1, Laura Monti1, Marta Lualdi1, Marco Fabbri2, Maria Grazia Sacco2, Laura Gribaldo2, and Roberto Taramelli1 1 Dipartimento di Biotecnologie e Scienze Molecolari, Università degli Studi dell’Insubria, via JH Dunant 3, 21100 Varese, Italy 2 European Commission - Joint Research Centre Institute for Health and Consumer Protection Molecular Biology and Genomics unit TP 464, Via E. Fermi, 2749 21027 Ispra (VA) - Italy Correspondence to: Roberto Taramelli, email: [email protected] Correspondence to: Francesco Acquati, email: [email protected] Keywords: RNases, cancer microenvironment, transcriptional profile Received: May 10, 2011, Accepted: June 2, 2011, Published: June 4, 2011 Copyright: © Acquati et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT: Using the Hey3Met2 human ovarian cancer cell line, we previously found the RNASET2 gene to possess a remarkable in vivo tumor suppressor activity, although no in vitro features such as inhibition of cell proliferation, clonogenic potential, impaired growth in soft agar and increase in apoptotic rate could be detected. This is reminiscent of the behavior of genes belonging to the class of tumor antagonizing genes (TAG) which act mainly within the context of the microenvironment. Here we present transcriptional profiles analysis which indicates that investigations of the mechanisms of TAG biological functions require a comparison between the in vitro and in vivo expression patterns. -

Towards an Understanding of Regulating Cajal Body Activity by Protein Modification
RNA BIOLOGY 2017, VOL. 14, NO. 6, 761–778 https://doi.org/10.1080/15476286.2016.1243649 REVIEW Towards an understanding of regulating Cajal body activity by protein modification Michael D. Hebert and Aaron R. Poole Department of Biochemistry, The University of Mississippi Medical Center, Jackson, MS, USA ABSTRACT ARTICLE HISTORY The biogenesis of small nuclear ribonucleoproteins (snRNPs), small Cajal body-specific RNPs (scaRNPs), Received 31 May 2016 small nucleolar RNPs (snoRNPs) and the telomerase RNP involves Cajal bodies (CBs). Although many Revised 30 August 2016 components enriched in the CB contain post-translational modifications (PTMs), little is known about how Accepted 27 September 2016 these modifications impact individual protein function within the CB and, in concert with other modified KEYWORDS factors, collectively regulate CB activity. Since all components of the CB also reside in other cellular Cajal body; coilin; locations, it is also important that we understand how PTMs affect the subcellular localization of CB phosphorylation; post- components. In this review, we explore the current knowledge of PTMs on the activity of proteins known translational modification; to enrich in CBs in an effort to highlight current progress as well as illuminate paths for future SMN; telomerase; WRAP53 investigation. Introduction functions of these proteins in the CB is diverse, but collectively There are different types of ribonucleoproteins (RNPs), which are thought to contribute to the RNP biogenesis mission of the are comprised of non-coding RNA and associated proteins. CB. Like other cellular processes, RNP biogenesis, and thus CB RNPs take part in fundamental cellular activities such as trans- activity, is regulated but an understanding of this regulation is lation and pre-mRNA splicing. -

Detailed Characterization of Human Induced Pluripotent Stem Cells Manufactured for Therapeutic Applications
Stem Cell Rev and Rep DOI 10.1007/s12015-016-9662-8 Detailed Characterization of Human Induced Pluripotent Stem Cells Manufactured for Therapeutic Applications Behnam Ahmadian Baghbaderani 1 & Adhikarla Syama2 & Renuka Sivapatham3 & Ying Pei4 & Odity Mukherjee2 & Thomas Fellner1 & Xianmin Zeng3,4 & Mahendra S. Rao5,6 # The Author(s) 2016. This article is published with open access at Springerlink.com Abstract We have recently described manufacturing of hu- help determine which set of tests will be most useful in mon- man induced pluripotent stem cells (iPSC) master cell banks itoring the cells and establishing criteria for discarding a line. (MCB) generated by a clinically compliant process using cord blood as a starting material (Baghbaderani et al. in Stem Cell Keywords Induced pluripotent stem cells . Embryonic stem Reports, 5(4), 647–659, 2015). In this manuscript, we de- cells . Manufacturing . cGMP . Consent . Markers scribe the detailed characterization of the two iPSC clones generated using this process, including whole genome se- quencing (WGS), microarray, and comparative genomic hy- Introduction bridization (aCGH) single nucleotide polymorphism (SNP) analysis. We compare their profiles with a proposed calibra- Induced pluripotent stem cells (iPSCs) are akin to embryonic tion material and with a reporter subclone and lines made by a stem cells (ESC) [2] in their developmental potential, but dif- similar process from different donors. We believe that iPSCs fer from ESC in the starting cell used and the requirement of a are likely to be used to make multiple clinical products. We set of proteins to induce pluripotency [3]. Although function- further believe that the lines used as input material will be used ally identical, iPSCs may differ from ESC in subtle ways, at different sites and, given their immortal status, will be used including in their epigenetic profile, exposure to the environ- for many years or even decades. -
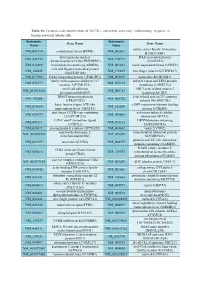
Systematic Name Gene Name Systematic Name Gene Name NM 001710 Complement Factor B(CFB) NM 052831 Solute Carrier Family 18 Member
Table S1: Genome-wide identification of SGLT2i`s interaction with early inflammatory response in human proximal tubular cells. Systematic Systematic Gene Name Gene Name Name Name solute carrier family 18 member NM_001710 complement factor B(CFB) NM_052831 B1(SLC18B1) heterogeneous nuclear DAZ associated protein NM_031372 NM_170711 ribonucleoprotein D like(HNRNPDL) 1(DAZAP1) NM_014299 bromodomain containing 4(BRD4) NM_001261 cyclin dependent kinase 9(CDK9) cilia and flagella associated protein NM_182628 NM_178835 zinc finger protein 827(ZNF827) 100(CFAP100) NM_017906 PAK1 interacting protein 1(PAK1IP1) NM_024015 homeobox B4(HOXB4) family with sequence similarity 167 ankyrin repeat and LEM domain NM_053279 NM_015114 member A(FAM167A) containing 2(ANKLE2) small cell adhesion ARP3 actin related protein 3 NM_001031628 NM_005721 glycoprotein(SMAGP) homolog(ACTR3) TRAF3 interacting protein actin related protein 2/3 complex NM_147686 NM_005720 2(TRAF3IP2) subunit 1B(ARPC1B) basic leucine zipper ATF-like cAMP responsive element binding NM_018664 NM_182898 transcription factor 3(BATF3) protein 5(CREB5) zinc finger CCCH-type containing activation induced cytidine NM_025079 NM_020661 12A(ZC3H12A) deaminase(AICDA) C-X-C motif chemokine ligand DENN domain containing NM_001511 NM_015213 1(CXCL1) 5A(DENND5A) NM_025072 prostaglandin E synthase 2(PTGES2) NM_004665 vanin 2(VNN2) superoxide dismutase 2, mitochondrial ribosomal protein NM_001024465 NM_016070 mitochondrial(SOD2) S23(MRPS23) jumonji and AT-rich interaction NM_033199 urocortin 2(UCN2) NM_004973 -

Therapeutic Vaccines for Amyotrophic Lateral Sclerosis Directed Against
Vaccine 37 (2019) 4920–4927 Contents lists available at ScienceDirect Vaccine journal homepage: www.elsevier.com/locate/vaccine Therapeutic vaccines for amyotrophic lateral sclerosis directed against disease specific epitopes of superoxide dismutase 1 ⇑ Beibei Zhao a, Kristen Marciniuk b,c, Ebrima Gibbs a, Masoud Yousefi a, Scott Napper b,c, Neil R. Cashman a, a Djavad Mowafaghian Centre for Brain Health, University of British Columbia, Vancouver, BC V6T 2B5, Canada b Department of Biochemistry, Microbiology, and Immunology, University of Saskatchewan, Saskatoon, Saskatchewan S7N 5E5, Canada c Vaccine and Infectious Disease Organization International Vaccine Research Center, University of Saskatchewan, Saskatchewan, SK S7N 5E3, Canada article info abstract Article history: Emerging evidence suggests seeding and prion-like propagation of mutant Superoxide Dismutase 1 Received 6 February 2019 (SOD1) misfolding to be a potential mechanism for ALS pathogenesis and progression. Immuno- Received in revised form 8 June 2019 targeting of misfolded SOD1 has shown positive clinical outcomes in mutant SOD1 transgenic mice. Accepted 10 July 2019 However, a major challenge in developing active immunotherapies for proteinopathies such as ALS is Available online 16 July 2019 the design of immunogens enabling exclusive recognition of pathogenic species of a self-protein. Ideally, one would achieve a robust antibody response against the disease-misfolded protein while spar- ing the natively folded conformer to avoid inducing deleterious autoimmune complications, or inhibiting its normal function. Using a motor neuron disease mouse model expressing human SOD1-G37R, we herein report the immunogenicity and therapeutic efficacy of two ALS vaccines, tgG-DSE2lim and tgG- DSE5b, based on the notion that native SOD1 would undergo early unfolding in disease to present ‘‘dis- ease specific epitopes” (DSE). -

A Genome Wide Association Study of Fast Beta EEG in Families of European Ancestry
CORE Metadata, citation and similar papers at core.ac.uk Provided by IUPUIScholarWorks HHS Public Access Author manuscript Author ManuscriptAuthor Manuscript Author Int J Psychophysiol Manuscript Author . Author Manuscript Author manuscript; available in PMC 2018 May 01. Published in final edited form as: Int J Psychophysiol. 2017 May ; 115: 74–85. doi:10.1016/j.ijpsycho.2016.12.008. A Genome Wide Association Study of Fast Beta EEG in Families of European Ancestry Jacquelyn L. Meyers1,*, Jian Zhang1, Niklas Manz1,2, Madhavi Rangaswamy3, Chella Kamarajan1, Leah Wetherill4, David B. Chorlian1, Sun J. Kang5, Lance Bauer6, Victor Hesselbrock6, John Kramer7, Samuel Kuperman7, John I. Nurnberger Jr4, Jay Tischfield8, Jen Chyong Wang9, Howard J. Edenberg4,10, Alison Goate9, Tatiana Foroud4, and Bernice Porjesz1 1Henri Begleiter Neurodynamics Laboratory, Department of Psychiatry and Behavioral Sciences, SUNY Downstate Medical Center, Brooklyn, NY, USA 2Department of Physics, College of Wooster, Wooster, OH, USA 3Department of Psychology, Christ University, Bangalore, India 4Department of Medical and Molecular Genetics, Indiana University School of Medicine, Indianapolis, IN, USA 5Albany Stratton VA Medical Center, Albany, NY, USA 6University of Connecticut School of Medicine, Farmington, CT, USA 7Department of Psychiatry, University of Iowa, Iowa City, IA, USA 8Rutgers University, Piscataway, NJ, USA 9Icahn School of Medicine at Mt. Sinai, New York, NY, USA 10Department of Biochemistry and Molecular Biology, Indiana University School of Medicine, Indianapolis, IN, USA Abstract BACKGROUND—Differences in fast beta (20–28 Hz) electroencephalogram (EEG) oscillatory activity distinguish some individuals with psychiatric and substance use disorders, suggesting that it may be a useful endophenotype for studying the genetics of disorders characterized by neural hyper-excitability. -

A Genomic Approach to Delineating the Occurrence of Scoliosis in Arthrogryposis Multiplex Congenita
G C A T T A C G G C A T genes Article A Genomic Approach to Delineating the Occurrence of Scoliosis in Arthrogryposis Multiplex Congenita Xenia Latypova 1, Stefan Giovanni Creadore 2, Noémi Dahan-Oliel 3,4, Anxhela Gjyshi Gustafson 2, Steven Wei-Hung Hwang 5, Tanya Bedard 6, Kamran Shazand 2, Harold J. P. van Bosse 5 , Philip F. Giampietro 7,* and Klaus Dieterich 8,* 1 Grenoble Institut Neurosciences, Université Grenoble Alpes, Inserm, U1216, CHU Grenoble Alpes, 38000 Grenoble, France; [email protected] 2 Shriners Hospitals for Children Headquarters, Tampa, FL 33607, USA; [email protected] (S.G.C.); [email protected] (A.G.G.); [email protected] (K.S.) 3 Shriners Hospitals for Children, Montreal, QC H4A 0A9, Canada; [email protected] 4 School of Physical & Occupational Therapy, Faculty of Medicine and Health Sciences, McGill University, Montreal, QC H3G 2M1, Canada 5 Shriners Hospitals for Children, Philadelphia, PA 19140, USA; [email protected] (S.W.-H.H.); [email protected] (H.J.P.v.B.) 6 Alberta Congenital Anomalies Surveillance System, Alberta Health Services, Edmonton, AB T5J 3E4, Canada; [email protected] 7 Department of Pediatrics, University of Illinois-Chicago, Chicago, IL 60607, USA 8 Institut of Advanced Biosciences, Université Grenoble Alpes, Inserm, U1209, CHU Grenoble Alpes, 38000 Grenoble, France * Correspondence: [email protected] (P.F.G.); [email protected] (K.D.) Citation: Latypova, X.; Creadore, S.G.; Dahan-Oliel, N.; Gustafson, Abstract: Arthrogryposis multiplex congenita (AMC) describes a group of conditions characterized A.G.; Wei-Hung Hwang, S.; Bedard, by the presence of non-progressive congenital contractures in multiple body areas. -

Downloaded Per Proteome Cohort Via the Web- Site Links of Table 1, Also Providing Information on the Deposited Spectral Datasets
www.nature.com/scientificreports OPEN Assessment of a complete and classifed platelet proteome from genome‑wide transcripts of human platelets and megakaryocytes covering platelet functions Jingnan Huang1,2*, Frauke Swieringa1,2,9, Fiorella A. Solari2,9, Isabella Provenzale1, Luigi Grassi3, Ilaria De Simone1, Constance C. F. M. J. Baaten1,4, Rachel Cavill5, Albert Sickmann2,6,7,9, Mattia Frontini3,8,9 & Johan W. M. Heemskerk1,9* Novel platelet and megakaryocyte transcriptome analysis allows prediction of the full or theoretical proteome of a representative human platelet. Here, we integrated the established platelet proteomes from six cohorts of healthy subjects, encompassing 5.2 k proteins, with two novel genome‑wide transcriptomes (57.8 k mRNAs). For 14.8 k protein‑coding transcripts, we assigned the proteins to 21 UniProt‑based classes, based on their preferential intracellular localization and presumed function. This classifed transcriptome‑proteome profle of platelets revealed: (i) Absence of 37.2 k genome‑ wide transcripts. (ii) High quantitative similarity of platelet and megakaryocyte transcriptomes (R = 0.75) for 14.8 k protein‑coding genes, but not for 3.8 k RNA genes or 1.9 k pseudogenes (R = 0.43–0.54), suggesting redistribution of mRNAs upon platelet shedding from megakaryocytes. (iii) Copy numbers of 3.5 k proteins that were restricted in size by the corresponding transcript levels (iv) Near complete coverage of identifed proteins in the relevant transcriptome (log2fpkm > 0.20) except for plasma‑derived secretory proteins, pointing to adhesion and uptake of such proteins. (v) Underrepresentation in the identifed proteome of nuclear‑related, membrane and signaling proteins, as well proteins with low‑level transcripts. -

Content Based Search in Gene Expression Databases and a Meta-Analysis of Host Responses to Infection
Content Based Search in Gene Expression Databases and a Meta-analysis of Host Responses to Infection A Thesis Submitted to the Faculty of Drexel University by Francis X. Bell in partial fulfillment of the requirements for the degree of Doctor of Philosophy November 2015 c Copyright 2015 Francis X. Bell. All Rights Reserved. ii Acknowledgments I would like to acknowledge and thank my advisor, Dr. Ahmet Sacan. Without his advice, support, and patience I would not have been able to accomplish all that I have. I would also like to thank my committee members and the Biomed Faculty that have guided me. I would like to give a special thanks for the members of the bioinformatics lab, in particular the members of the Sacan lab: Rehman Qureshi, Daisy Heng Yang, April Chunyu Zhao, and Yiqian Zhou. Thank you for creating a pleasant and friendly environment in the lab. I give the members of my family my sincerest gratitude for all that they have done for me. I cannot begin to repay my parents for their sacrifices. I am eternally grateful for everything they have done. The support of my sisters and their encouragement gave me the strength to persevere to the end. iii Table of Contents LIST OF TABLES.......................................................................... vii LIST OF FIGURES ........................................................................ xiv ABSTRACT ................................................................................ xvii 1. A BRIEF INTRODUCTION TO GENE EXPRESSION............................. 1 1.1 Central Dogma of Molecular Biology........................................... 1 1.1.1 Basic Transfers .......................................................... 1 1.1.2 Uncommon Transfers ................................................... 3 1.2 Gene Expression ................................................................. 4 1.2.1 Estimating Gene Expression ............................................ 4 1.2.2 DNA Microarrays ...................................................... -

Spliceosomal Usnrnp Biogenesis, Structure and Function Cindy L Will* and Reinhard Lührmann†
290 Spliceosomal UsnRNP biogenesis, structure and function Cindy L Will* and Reinhard Lührmann† Significant advances have been made in elucidating the for each of the two reaction steps. Components of the biogenesis pathway and three-dimensional structure of the UsnRNPs also appear to catalyze the two transesterifi- UsnRNPs, the building blocks of the spliceosome. U2 and cation reactions leading to excision of the intron and U4/U6•U5 tri-snRNPs functionally associate with the pre-mRNA ligation of the 5′ and 3′ exons. Here we describe recent at an earlier stage of spliceosome assembly than previously advances in our understanding of snRNP biogenesis, thought, and additional evidence supporting UsnRNA-mediated structure and function, focusing primarily on the major catalysis of pre-mRNA splicing has been presented. spliceosomal UsnRNPs from higher eukaryotes. Addresses Identification of a novel UsnRNA export factor Max Planck Institute of Biophysical Chemistry, Department of Cellular UsnRNP biogenesis is a complex process, many aspects of Biochemistry, Am Fassberg 11, 37077 Göttingen, Germany. which remain poorly understood. Although less is known *e-mail: [email protected] about the maturation process of the minor U11, U12 and †e-mail: [email protected] U4atac UsnRNPs, it is assumed that they follow a pathway Current Opinion in Cell Biology 2001, 13:290–301 similar to that described below for the major UsnRNPs 0955-0674/01/$ — see front matter (see Figure 1). The UsnRNAs, with the exception of U6 © 2001 Elsevier Science Ltd. All rights reserved. and U6atac (see below), are transcribed by RNA polymerase II as snRNA precursors that contain additional Abbreviations ′ CBC cap-binding complex 3 nucleotides and acquire a monomethylated, m7GpppG NLS nuclear localization signal (m7G) cap structure. -

Lifestyle Intervention in Pregnant Women with Obesity Impacts Cord Blood DNA Methylation, Which Associates with Body Composition in the Offspring
854 Diabetes Volume 70, April 2021 Lifestyle Intervention in Pregnant Women With Obesity Impacts Cord Blood DNA Methylation, Which Associates With Body Composition in the Offspring Josefine Jönsson,1 Kristina M. Renault,2,3 Sonia García-Calzón,1,4 Alexander Perfilyev,1 Angela C. Estampador,5 Kirsten Nørgaard,6 Mads Vendelbo Lind,7 Allan Vaag,6 Line Hjort,8 Kim F. Michaelsen,7 Emma Malchau Carlsen,7,9 Paul W. Franks,5 and Charlotte Ling1 Diabetes 2021;70:854–866 | https://doi.org/10.2337/db20-0487 Maternal obesity may lead to epigenetic alterations in the mediates the effect of the lifestyle intervention on lean offspring and might thereby contribute to disease later in mass in the offspring (FDR <5%). Moreover, 22 methyla- life. We investigated whether a lifestyle intervention in tion sites were associated with offspring BMI z scores pregnant women with obesity is associated with epige- during the first 3 years of life (P < 0.05). Overall, lifestyle netic variation in cord blood and body composition in the interventions in pregnant women with obesity are asso- offspring. Genome-wide DNA methylation was analyzed ciated with epigenetic changes in offspring, potentially in cord blood from 208 offspring from the Treatment of influencing the offspring’s lean mass and early growth. Obese Pregnant women (TOP)-study, which includes pregnant women with obesity randomized to lifestyle interventions comprised of physical activity with or with- Obesity and type 2 diabetes are on the rise worldwide, as is out dietary advice versus control subjects (standard of the prevalence of obesity in pregnant women (1).