PDF-Document
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Heat Resistant Thermophilic Endospores in Cold Estuarine Sediments
Heat resistant thermophilic endospores in cold estuarine sediments Emma Bell Thesis submitted for the degree of Doctor of Philosophy School of Civil Engineering and Geosciences Faculty of Science, Agriculture and Engineering February 2016 Abstract Microbial biogeography explores the spatial and temporal distribution of microorganisms at multiple scales and is influenced by environmental selection and passive dispersal. Understanding the relative contribution of these factors can be challenging as their effects can be difficult to differentiate. Dormant thermophilic endospores in cold sediments offer a natural model for studies focusing on passive dispersal. Understanding distributions of these endospores is not confounded by the influence of environmental selection; rather their occurrence is due exclusively to passive transport. Sediment heating experiments were designed to investigate the dispersal histories of various thermophilic spore-forming Firmicutes in the River Tyne, a tidal estuary in North East England linking inland tributaries with the North Sea. Microcosm incubations at 50-80°C were monitored for sulfate reduction and enriched bacterial populations were characterised using denaturing gradient gel electrophoresis, functional gene clone libraries and high-throughput sequencing. The distribution of thermophilic endospores among different locations along the estuary was spatially variable, indicating that dispersal vectors originating in both warm terrestrial and marine habitats contribute to microbial diversity in estuarine and marine environments. In addition to their persistence in cold sediments, some endospores displayed a remarkable heat-resistance surviving multiple rounds of autoclaving. These extremely heat-resistant endospores are genetically similar to those detected in deep subsurface environments, including geothermal groundwater investigated from a nearby terrestrial borehole drilled to >1800 m depth with bottom temperatures in excess of 70°C. -

WO 2018/064165 A2 (.Pdf)
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2018/064165 A2 05 April 2018 (05.04.2018) W !P O PCT (51) International Patent Classification: Published: A61K 35/74 (20 15.0 1) C12N 1/21 (2006 .01) — without international search report and to be republished (21) International Application Number: upon receipt of that report (Rule 48.2(g)) PCT/US2017/053717 — with sequence listing part of description (Rule 5.2(a)) (22) International Filing Date: 27 September 2017 (27.09.2017) (25) Filing Language: English (26) Publication Langi English (30) Priority Data: 62/400,372 27 September 2016 (27.09.2016) US 62/508,885 19 May 2017 (19.05.2017) US 62/557,566 12 September 2017 (12.09.2017) US (71) Applicant: BOARD OF REGENTS, THE UNIVERSI¬ TY OF TEXAS SYSTEM [US/US]; 210 West 7th St., Austin, TX 78701 (US). (72) Inventors: WARGO, Jennifer; 1814 Bissonnet St., Hous ton, TX 77005 (US). GOPALAKRISHNAN, Vanch- eswaran; 7900 Cambridge, Apt. 10-lb, Houston, TX 77054 (US). (74) Agent: BYRD, Marshall, P.; Parker Highlander PLLC, 1120 S. Capital Of Texas Highway, Bldg. One, Suite 200, Austin, TX 78746 (US). (81) Designated States (unless otherwise indicated, for every kind of national protection available): AE, AG, AL, AM, AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, HR, HU, ID, IL, IN, IR, IS, JO, JP, KE, KG, KH, KN, KP, KR, KW, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. -
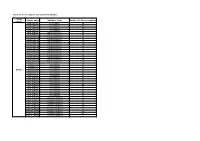
Table S1: List of Samples Included in the Analysis
Table S1: list of samples included in the analysis Study Sample name Inhibitory status Number of days at sampling number DNA.0P2T4 No inhibition 29 DNA.0P2T6 No inhibition 57 DNA.10P2T4 No inhibition 29 DNA.10P2T6 No inhibition 57 DNA.75P2T4 Phenol inhibition 29 DNA.75P2T6 Phenol inhibition 57 DNA.100P2T4 Phenol inhibition 29 DNA.100P2T6 Phenol inhibition 57 DNA.125P1T4 Phenol inhibition 29 DNA.125P1T6 Phenol inhibition 57 DNA.125P2T4 Phenol inhibition 29 DNA.125P2T6 Phenol inhibition 57 DNA.125P3T4 Phenol inhibition 29 DNA.125P3T6 Phenol inhibition 57 DNA.150P2T4 Phenol inhibition 29 DNA.150P2T6 Phenol inhibition 57 DNA.200P2T4 Phenol inhibition 29 DNA.200P2T6 Phenol inhibition 57 DNA.0N2T4 No inhibition 29 DNA.0N2T5 No inhibition 42 DNA.0N2T6 No inhibition 57 Study 1 DNA.5N2T4 No inhibition 29 DNA.5N2T5 No inhibition 42 DNA.5N2T6 No inhibition 57 DNA.10N2T4 No inhibition 29 DNA.10N2T5 No inhibition 42 DNA.10N2T6 No inhibition 57 DNA.15N2T4 No inhibition 29 DNA.15N2T5 No inhibition 42 DNA.15N2T6 No inhibition 57 DNA.25N2T4 No inhibition 29 DNA.25N2T5 No inhibition 42 DNA.25N2T6 No inhibition 57 DNA.75N2T4 Ammonia inhibition 29 DNA.75N2T5 Ammonia inhibition 42 DNA.75N2T6 Ammonia inhibition 57 DNA.100N2T4 Ammonia inhibition 29 DNA.100N2T5 Ammonia inhibition 42 DNA.100N2T6 Ammonia inhibition 57 DNA.250N2T4 Ammonia inhibition 29 DNA.250N2T5 Ammonia inhibition 42 DNA.250N2T6 Ammonia inhibition 57 nono2T3 No inhibition 16 noN2T4 Ammonia inhibition 23 noN2T8 Ammonia inhibition 60 noN2T9 Ammonia inhibition 85 noPhi2T4 Phenol inhibition 23 noPhi2T5 -

Physiologica Thermoa Al and Phylogenetic Stu Caloramator And
University of Akureyri Faculty of Business and Science Department of Natural Resource Sciences Physiological and phylogenetic studies of Caloramator and Thermoanaerobacterium species: Ethanol and hydrogen production from complex biomass Margrét Auður Sigurbjörnsdóttir Supervisor: Jóhann Örlygsson University of Akureyri Submitted in part fulfillment of the degree of Master of Science in Natural Resource Science – Biotechnology May 2009 University of Akureyri Department of Business and Science “There are in fact two things, science and opinion; the former begets knowledge, the latter ignorance.” Hippocrates ii University of Akureyri Department of Business and Science University of Akureyri Faculty of Business and Science Department of Natural Resource Sciences Physiological and phylogenetic studies of Caloramator and Thermoanaerobacterium species: Ethanol and hydrogen production from complex biomass Margrét Auður Sigurbjörnsdóttir Master thesis for 90 credit M.Sc. in Biotechnology Printed copies: 10 Number of pages: 109 Appendixes: 2 iii University of Akureyri Department of Business and Science Declaration I hereby declare that I am the only author of this thesis and it is the product of my own research. _______________________________________________ Margrét Auður Sigurbjörnsdóttir It is hereby confirmed that this master thesis is satisfactory to M.Sc. – degree from the Faculty of Business and Science, department of Natural Resource Science. ________________________________________________ Dr. Jóhann Örlygsson iv University of Akureyri Department of Business and Science Abstract Three anerobic, thermophilic bacteria were isolated from hot springs in Graendalur, SW-Iceland. The strains were investigated with respect to phylogenetic, physiology and end product formation. Phylogenetic studies were done with both partial and full 16S rRNA sequence analysis on all three strains. Two of the strains, AK44 and AK35, belong to the genus Caloramator with the closest relation to C. -

Genome Diversity of Spore-Forming Firmicutes MICHAEL Y
Genome Diversity of Spore-Forming Firmicutes MICHAEL Y. GALPERIN National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20894 ABSTRACT Formation of heat-resistant endospores is a specific Vibrio subtilis (and also Vibrio bacillus), Ferdinand Cohn property of the members of the phylum Firmicutes (low-G+C assigned it to the genus Bacillus and family Bacillaceae, Gram-positive bacteria). It is found in representatives of four specifically noting the existence of heat-sensitive vegeta- different classes of Firmicutes, Bacilli, Clostridia, Erysipelotrichia, tive cells and heat-resistant endospores (see reference 1). and Negativicutes, which all encode similar sets of core sporulation fi proteins. Each of these classes also includes non-spore-forming Soon after that, Robert Koch identi ed Bacillus anthracis organisms that sometimes belong to the same genus or even as the causative agent of anthrax in cattle and the species as their spore-forming relatives. This chapter reviews the endospores as a means of the propagation of this orga- diversity of the members of phylum Firmicutes, its current taxon- nism among its hosts. In subsequent studies, the ability to omy, and the status of genome-sequencing projects for various form endospores, the specific purple staining by crystal subgroups within the phylum. It also discusses the evolution of the violet-iodine (Gram-positive staining, reflecting the pres- Firmicutes from their apparently spore-forming common ancestor ence of a thick peptidoglycan layer and the absence of and the independent loss of sporulation genes in several different lineages (staphylococci, streptococci, listeria, lactobacilli, an outer membrane), and the relatively low (typically ruminococci) in the course of their adaptation to the saprophytic less than 50%) molar fraction of guanine and cytosine lifestyle in a nutrient-rich environment. -

Community Analysis of Plant Biomass-Degrading Microorganisms from Obsidian Pool, Yellowstone National Park
Microb Ecol DOI 10.1007/s00248-014-0500-8 ENVIRONMENTAL MICROBIOLOGY Community Analysis of Plant Biomass-Degrading Microorganisms from Obsidian Pool, Yellowstone National Park Tatiana A. Vishnivetskaya & Scott D. Hamilton-Brehm & Mircea Podar & Jennifer J. Mosher & Anthony V. Palumbo & Tommy J. Phelps & Martin Keller & James G. Elkins Received: 12 May 2014 /Accepted: 16 September 2014 # Springer Science+Business Media New York (outside the USA) 2014 Abstract The conversion of lignocellulosic biomass into (OBP), was examined for potential biomass-active microorgan- biofuels can potentially be improved by employing robust mi- isms using cultivation-independent and enrichment techniques. croorganisms and enzymes that efficiently deconstruct plant Analysis of 33,684 archaeal and 43,784 bacterial quality-filtered polysaccharides at elevated temperatures. Many of the geother- 16S rRNA gene pyrosequences revealed that archaeal diversity mal features of Yellowstone National Park (YNP) are surrounded in the main pool was higher than bacterial; however, in the by vegetation providing a source of allochthonic material to vegetated area, overall bacterial diversity was significantly support heterotrophic microbial communities adapted to utilize higher. Of notable interest was a flooded depression adjacent to plant biomass as a primary carbon and energy source. In this OBP supporting a stand of Juncus tweedyi, a heat-tolerant rush study, a well-known hot spring environment, Obsidian Pool commonly found growing near geothermal features in YNP. The microbial community from heated sediments surrounding the The submitted manuscript has been authored by a contractor of the U.S. plants was enriched in members of the Firmicutes including Government under contract DE-AC05-00OR22725. Accordingly, the potentially (hemi)cellulolytic bacteria from the genera U.S. -

Functional Comparison of Bacteria from the Human Gut and Closely
Functional comparison of bacteria from the human gut and closely related non-gut bacteria reveals the importance of conjugation and a paucity of motility and chemotaxis functions in the gut environment Dragana Dobrijevic, Anne-Laure Abraham, Alexandre Jamet, Emmanuelle Maguin, Maarten van de Guchte To cite this version: Dragana Dobrijevic, Anne-Laure Abraham, Alexandre Jamet, Emmanuelle Maguin, Maarten van de Guchte. Functional comparison of bacteria from the human gut and closely related non-gut bacte- ria reveals the importance of conjugation and a paucity of motility and chemotaxis functions in the gut environment. PLoS ONE, Public Library of Science, 2016, 11 (7), pp.e0159030. 10.1371/jour- nal.pone.0159030. hal-01353535 HAL Id: hal-01353535 https://hal.archives-ouvertes.fr/hal-01353535 Submitted on 11 Aug 2016 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License RESEARCH ARTICLE Functional Comparison of Bacteria from the Human Gut and Closely Related Non-Gut Bacteria Reveals -

Anaerobic Digestion of Biofuel Production Residues
ANAEROBIC DIGESTION OF BIOFUEL PRODUCTION RESIDUES By ZHUOLI TIAN A DISSERTATION PRESENTED TO THE GRADUATE SCHOOL OF THE UNIVERSITY OF FLORIDA IN PARTIAL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY UNIVERSITY OF FLORIDA 2011 1 © 2011 Zhuoli Tian 2 To my parents who have always been supportive of my education 3 ACKNOWLEDGMENTS I would like to extend my deepest appreciation to my parents, Mr.Tian and Mrs. Zheng for their continuous support and encouragement towards my academic endeavors. I express my sincere gratitude to my advisor and committee chairman, Dr. Pratap Pullammanappallil, for his continual support and guidance during my time at the University of Florida. I am grateful to Dr. Lonnie O. Ingram for his support throughout this research study. I would like to thank Dr. Ben Koopman and Dr. Paul A. Chadik for serving on my committee and their inspirational lectures. I also own gratitude to Dr. Melanie Correll for her devotion and patience in helping improve my writing skills. I give thanks to Dr. Adriana Giongo and Ms. Diane Chauliac for their guidance and assistance in carrying out microbial anaysis for this research. I also would like to thank Dr. Claudia C. Geddes and Dr. Mike Mullinnix for providing necessary materials and data for this research, as well as their insightful ideas and assistances in solving analytical issues. I would like to thank the following individuals for their supports and assistances during my studies, Abhay Koppar, Cesar Moreira , Congrong Yu, Gayathri Ram Mohan, Ioannis M. Polematidis, Jaime Chavez Leon, Mandu Inyang, Patrick Dube, Samriddhi Buxy, Yuan Tian and all the friends in the Department of Agricultural and Biological Engineering. -

Caloramator Proteoclasticus Sp. Nov., a New Moderately Thermophilic Anaerobic Proteolytic Bacterium
INTERNATIONALJOURNAL OF SYSTEMATICBACTERIOLOGY, July 1997, p. 651-656 Vol. 47, No. 3 0020-7713/97/$04.00+ 0 Copyright 0 1997, International Union of Microbiological Societies Caloramator proteoclasticus sp. nov., a New Moderately Thermophilic Anaerobic Proteolytic Bacterium SILVANA TARLERA,~"LUC~A MUXI,' MATILDE SOUBES,~AND ALFONS J. M. STAMS* Departamento de Microbiologia, Facultad de Quimica y Facultad de Ciencias, Universidad de la Republica, Montevideo, Uruguay, ' and Department of Microbiology, Wageningen Agricultural University, 6703 CT Wageningen, The Netherlands2 A new moderately thermophilic proteolytic anaerobe, strain UT, was isolated from mesophilic granular methanogenic sludge. The cells were spore-forming, motile rods that were 0.4 pm wide and 2.4 to 4 pm long and stained gram negative. Electron micrographs of thin sections revealed the presence of an atypical gram-positive cell wall. Optimum growth occurred at 55°C and at pH values between 7.0 and 7.5, with a doubling time of 30 min. The DNA base ratio of guanine plus cytosine was 31 mol%. The bacterium fermented proteins mainly to acetate, hydrogen, formate, and branched-chain fatty acids. Several amino acids, including glutamate, aspartate, arginine, histidine, threonine, methionine, and branched-chain amino acids, were also utilized. Glutamate was degraded to acetate, formate, hydrogen, and alanine. In addition, the strain degraded carbohydrates, including glucose, fructose, mannose, cellobiose, and starch, to acetate, ethanol, formate, lactate, and hydrogen. The results of a 16s rRNA sequence analysis phylogenetically placed strain UT in the low-guanine-plus-cytosine-contentsubgroup of the gram-positive phylum. We propose to classify the described strain in the genus Caloramator as a new species, Caloramator proteoclasticus. -

Large Scale Function-Based Genome Prospecting for Industrial Traits Applied to 1,3-Propanediol Production
bioRxiv preprint doi: https://doi.org/10.1101/2021.08.25.457110; this version posted August 27, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Large scale function-based genome prospecting for industrial traits applied to 1,3-propanediol production. Jasper J. Koehorst ID 1*, , Nikolaos Strepis ID 1,2*, Sanne de Graaf1, Alfons J. M. Stams ID 2,3, Diana Z. Sousa ID 2, and Peter J. Schaap ID 1 1Laboratory of Systems and Synthetic Biology | Department of Agrotechnology and Food Sciences, Wageningen University and Research, Wageningen, the Netherlands 2Laboratory of Microbiology | Department of Agrotechnology and Food Sciences, Wageningen University and Research, Wageningen, the Netherlands 3Centre of Biological Engineering, University of Minho, 4710-057 Braga, Portugal Due to the success of next-generation sequencing, there has been normally used. However, over larger phylogenetic distances, a vast build-up of microbial genomes in the public reposito- strategies that use sequence-based clustering algorithms are ries. FAIR genome prospecting of this huge genomic potential hampered by accumulation of evolutionary signals, lateral for biotechnological benefiting, require new efficient and flexible gene transfer, gene fusion/fission events and domain expan- methods. In this study, Semantic Web technologies are applied sions (5). Furthermore, when applied at larger scales, such to develop a function-based genome mining approach that fol- strategies suffer from high computational time and mem- lows a knowledge and discovery in database (KDD) protocol. -
Gut Microbiota and Fecal Metabolome Perturbation in Children with Autism Spectrum Disorder
DOI: 10.15171/mejdd.2018.112 Review Article 205 Gut Microbiota and Fecal Metabolome Perturbation in Children with Autism Spectrum Disorder Ashraf Mohamadkhani 1,* ABSTRACT 1. Liver and Pancreatobiliary Diseases The brain-intestinal axis concept describes the communication between the intestinal Research Center, Digestive Disease microbiota as an ecosystem of a number of dynamic microorganisms and the brain. The Research Institute, Shariati Hospital, composition of the microbial community of the human gut is important for human health Tehran University of Medical Sciences, by influencing the total metabolomic profile. In children with autism spectrum disorder Tehran, Iran (ASD), the composition of the fecal microbiota and their metabolic products has a different configuration of the healthy child. An imbalance in the metabolite derived from the microbiota in children with ASD affect brain development and social behavior. In this article, we review recent discoveries about intestinal metabolites derived from microbiota based on high-yield molecular studies in children with ASD as part of the “intestinal brain axis”. KEYWORDS: Autism spectrum disorder (ASD), Intestinal microbiota, Fecal metabolites, ASD children Please cite this paper as: Mohamadkhani A. Gut Microbiota and Fecal Metabolome Perturbation in Children with Autism Spectrum Disorder. Middle East J Dig Dis 2018;10:205-212. doi: 10.15171/ mejdd.2018.112. INTRODUCTION The development of intestinal microflora begins at birth and subse- quently the colonization of bacterial composition -

Literature Review: Thermal Environments and Biodiversity
GEOMICROBIOLOGICAL DESCRIPTION OF TWO CONTEMPORARY HYDROTHERMAL POOLS IN UZON CALDERA, KAMCHATKA, RUSSIA, AS MODELS FOR SULFUR BIOGEOCHEMISTRY by ELIZABETH ADRIENNE BURGESS (Under the Direction of Juergen Wiegel) ABSTRACT The combination of geological activity and geographic isolation make Uzon Caldera, Kamchatka, Russia, an ideal location for geomicrobiological research. Two hydrothermal pools, Arkashin Shurf (Arkashin) and Zavarzin Spring (Zavarzin), were selected for geochemical and microbiological characterization over multiple years and scales. Arkashin has high arsenic concentrations relative to Zavarzin, which has abundant elemental sulfur. Grab samples were analyzed from a geomicrobiological perspective, to describe community structure in each pool based on sequence, lipid and stable isotope data. Based on sequence analysis and lipid distribution, Arkashin was inhabited by Hydrogenobaculum- related primary producers. Other community members included Desulfurella-, ―Sphingobacteria―- and Variovorax-related microorganisms. Zavarzin was dominated by Chloroflexus, and heterotrophic microorganisms, including some Crenarchaeota. The community in Zavarzin was more diverse than in Arkashin. Additionally, nearly 20% of the sequences from Arkashin and over 50% of the sequences from Zavarzin represented uncultured and unclassified microorganisms. Core sample analyses indicated that in each pool the microbiology and geochemistry changed with depth over visible changes in color and texture. Surface sub-samples were similar to the grab samples. Autotrophic surface communities were replaced with microorganisms dependent on heterotrophic inputs or reduced hydrothermal carbon as depth increased. In Arkashin, variation in color of strata was associated with varying concentrations of As and S. The highest As and S concentrations were associated with the lowest concentrations of phospholipid fatty acids (PLFA). In Zavarzin, the highest sulfur and PLFA concentrations were associated with fine-textured surface samples.