SUPPLEMENTARY DATA Supplementary Table 1. Bacterial
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The 2014 Golden Gate National Parks Bioblitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event
National Park Service U.S. Department of the Interior Natural Resource Stewardship and Science The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 ON THIS PAGE Photograph of BioBlitz participants conducting data entry into iNaturalist. Photograph courtesy of the National Park Service. ON THE COVER Photograph of BioBlitz participants collecting aquatic species data in the Presidio of San Francisco. Photograph courtesy of National Park Service. The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 Elizabeth Edson1, Michelle O’Herron1, Alison Forrestel2, Daniel George3 1Golden Gate Parks Conservancy Building 201 Fort Mason San Francisco, CA 94129 2National Park Service. Golden Gate National Recreation Area Fort Cronkhite, Bldg. 1061 Sausalito, CA 94965 3National Park Service. San Francisco Bay Area Network Inventory & Monitoring Program Manager Fort Cronkhite, Bldg. 1063 Sausalito, CA 94965 March 2016 U.S. Department of the Interior National Park Service Natural Resource Stewardship and Science Fort Collins, Colorado The National Park Service, Natural Resource Stewardship and Science office in Fort Collins, Colorado, publishes a range of reports that address natural resource topics. These reports are of interest and applicability to a broad audience in the National Park Service and others in natural resource management, including scientists, conservation and environmental constituencies, and the public. The Natural Resource Report Series is used to disseminate comprehensive information and analysis about natural resources and related topics concerning lands managed by the National Park Service. -

Comparative Metagenomic Analysis of Microcosm Structures and Lignocellulolytic Enzyme Systems of Symbiotic Biomass-Degrading Consortia
Comparative metagenomic analysis of microcosm structures and lignocellulolytic enzyme systems of symbiotic biomass-degrading consortia Sarunyou Wongwilaiwalin & Thanaporn Laothanachareon & Wuttichai Mhuantong & Sithichoke Tangphatsornruang & Lily Eurwilaichitr & Yasuo Igarashi & Verawat Champreda Received: 2 October 2012 /Revised: 3 January 2013 /Accepted: 7 January 2013 # Springer-Verlag Berlin Heidelberg 2013 Abstract Decomposition of lignocelluloses by cooperative industrial pulp waste with CMCase, xylanase, and β- microbial actions is an essential process of carbon cycling in glucanase activities in the supernatant. Shotgun pyrosequenc- nature and provides a basis for biomass conversion to fuels ing of the BGC-1 metagenome indicated a markedly high and chemicals in biorefineries. In this study, structurally stable relative abundance of genes encoding for glycosyl hydrolases, symbiotic aero-tolerant lignocellulose-degrading microbial particularly for lignocellulytic enzymes in 26 families. The consortia were obtained from biodiversified microflora pres- enzyme system comprised a unique composition of main- ent in industrial sugarcane bagasse pile (BGC-1), cow rumen chain degrading and side-chain processing hydrolases, domi- fluid (CRC-1), and pulp mill activated sludge (ASC-1) by nated by GH2, 3, 5, 9, 10, and 43, reflecting adaptation of successive subcultivation on rice straw under facultative an- enzyme profiles to the specific substrate. Gene mapping oxic conditions. Tagged 16S rRNA gene pyrosequencing showed metabolic potential -

Fatty Acid Diets: Regulation of Gut Microbiota Composition and Obesity and Its Related Metabolic Dysbiosis
International Journal of Molecular Sciences Review Fatty Acid Diets: Regulation of Gut Microbiota Composition and Obesity and Its Related Metabolic Dysbiosis David Johane Machate 1, Priscila Silva Figueiredo 2 , Gabriela Marcelino 2 , Rita de Cássia Avellaneda Guimarães 2,*, Priscila Aiko Hiane 2 , Danielle Bogo 2, Verônica Assalin Zorgetto Pinheiro 2, Lincoln Carlos Silva de Oliveira 3 and Arnildo Pott 1 1 Graduate Program in Biotechnology and Biodiversity in the Central-West Region of Brazil, Federal University of Mato Grosso do Sul, Campo Grande 79079-900, Brazil; [email protected] (D.J.M.); [email protected] (A.P.) 2 Graduate Program in Health and Development in the Central-West Region of Brazil, Federal University of Mato Grosso do Sul, Campo Grande 79079-900, Brazil; pri.fi[email protected] (P.S.F.); [email protected] (G.M.); [email protected] (P.A.H.); [email protected] (D.B.); [email protected] (V.A.Z.P.) 3 Chemistry Institute, Federal University of Mato Grosso do Sul, Campo Grande 79079-900, Brazil; [email protected] * Correspondence: [email protected]; Tel.: +55-67-3345-7416 Received: 9 March 2020; Accepted: 27 March 2020; Published: 8 June 2020 Abstract: Long-term high-fat dietary intake plays a crucial role in the composition of gut microbiota in animal models and human subjects, which affect directly short-chain fatty acid (SCFA) production and host health. This review aims to highlight the interplay of fatty acid (FA) intake and gut microbiota composition and its interaction with hosts in health promotion and obesity prevention and its related metabolic dysbiosis. -

Effect of Ph and Temperature on Microbial Community Structure And
Calicioglu et al. Biotechnol Biofuels (2018) 11:275 https://doi.org/10.1186/s13068-018-1278-6 Biotechnology for Biofuels RESEARCH Open Access Efect of pH and temperature on microbial community structure and carboxylic acid yield during the acidogenic digestion of duckweed Ozgul Calicioglu1, Michael J. Shreve1, Tom L. Richard2 and Rachel A. Brennan1* Abstract Background: Duckweeds (Lemnaceae) are efcient aquatic plants for wastewater treatment due to their high nutri- ent-uptake capabilities and resilience to severe environmental conditions. Combined with their rapid growth rates, high starch, and low lignin contents, duckweeds have also gained popularity as a biofuel feedstock for thermochemi- cal conversion and alcohol fermentation. However, studies on the acidogenic anaerobic digestion of duckweed into carboxylic acids, another group of chemicals which are precursors of higher-value chemicals and biofuels, are lacking. In this study, a series of laboratory batch experiments were performed to determine the favorable operating condi- tions (i.e., temperature and pH) to maximize carboxylic acid production from wastewater-derived duckweed during acidogenic digestion. Batch reactors with 25 g/l solid loading were operated anaerobically for 21 days under meso- philic (35 °C) or thermophilic (55 °C) conditions at an acidic (5.3) or basic (9.2) pH. At the conclusion of the experiment, the dominant microbial communities under various operating conditions were assessed using high-throughput sequencing. Results: The highest duckweed–carboxylic acid conversion of 388 28 mg acetic acid equivalent per gram volatile solids was observed under mesophilic and basic conditions, with an± average production rate of 0.59 g/l/day. This result is comparable to those reported for acidogenic digestion of other organics such as food waste. -

Heat Resistant Thermophilic Endospores in Cold Estuarine Sediments
Heat resistant thermophilic endospores in cold estuarine sediments Emma Bell Thesis submitted for the degree of Doctor of Philosophy School of Civil Engineering and Geosciences Faculty of Science, Agriculture and Engineering February 2016 Abstract Microbial biogeography explores the spatial and temporal distribution of microorganisms at multiple scales and is influenced by environmental selection and passive dispersal. Understanding the relative contribution of these factors can be challenging as their effects can be difficult to differentiate. Dormant thermophilic endospores in cold sediments offer a natural model for studies focusing on passive dispersal. Understanding distributions of these endospores is not confounded by the influence of environmental selection; rather their occurrence is due exclusively to passive transport. Sediment heating experiments were designed to investigate the dispersal histories of various thermophilic spore-forming Firmicutes in the River Tyne, a tidal estuary in North East England linking inland tributaries with the North Sea. Microcosm incubations at 50-80°C were monitored for sulfate reduction and enriched bacterial populations were characterised using denaturing gradient gel electrophoresis, functional gene clone libraries and high-throughput sequencing. The distribution of thermophilic endospores among different locations along the estuary was spatially variable, indicating that dispersal vectors originating in both warm terrestrial and marine habitats contribute to microbial diversity in estuarine and marine environments. In addition to their persistence in cold sediments, some endospores displayed a remarkable heat-resistance surviving multiple rounds of autoclaving. These extremely heat-resistant endospores are genetically similar to those detected in deep subsurface environments, including geothermal groundwater investigated from a nearby terrestrial borehole drilled to >1800 m depth with bottom temperatures in excess of 70°C. -

WO 2018/064165 A2 (.Pdf)
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2018/064165 A2 05 April 2018 (05.04.2018) W !P O PCT (51) International Patent Classification: Published: A61K 35/74 (20 15.0 1) C12N 1/21 (2006 .01) — without international search report and to be republished (21) International Application Number: upon receipt of that report (Rule 48.2(g)) PCT/US2017/053717 — with sequence listing part of description (Rule 5.2(a)) (22) International Filing Date: 27 September 2017 (27.09.2017) (25) Filing Language: English (26) Publication Langi English (30) Priority Data: 62/400,372 27 September 2016 (27.09.2016) US 62/508,885 19 May 2017 (19.05.2017) US 62/557,566 12 September 2017 (12.09.2017) US (71) Applicant: BOARD OF REGENTS, THE UNIVERSI¬ TY OF TEXAS SYSTEM [US/US]; 210 West 7th St., Austin, TX 78701 (US). (72) Inventors: WARGO, Jennifer; 1814 Bissonnet St., Hous ton, TX 77005 (US). GOPALAKRISHNAN, Vanch- eswaran; 7900 Cambridge, Apt. 10-lb, Houston, TX 77054 (US). (74) Agent: BYRD, Marshall, P.; Parker Highlander PLLC, 1120 S. Capital Of Texas Highway, Bldg. One, Suite 200, Austin, TX 78746 (US). (81) Designated States (unless otherwise indicated, for every kind of national protection available): AE, AG, AL, AM, AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, HR, HU, ID, IL, IN, IR, IS, JO, JP, KE, KG, KH, KN, KP, KR, KW, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. -

Gut Microbiota Features Associated with Clostridioides Difficile Colonization in Dairy Calves
bioRxiv preprint doi: https://doi.org/10.1101/2021.05.11.443551; this version posted May 11, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 Gut microbiota features associated with Clostridioides 2 difficile colonization in dairy calves 3 4 Laurel E. Redding1, Alexander S. Berry2,3, Nagaraju Indugu1, Elizabeth Huang1, Daniel P. Beiting3, Dipti 5 Pitta1 6 7 8 1Department of Clinical Sciences, University of Pennsylvania, School of Veterinary Medicine, Kennett 9 Square, PA, USA 10 11 2Division of Gastroenterology, Hepatology, and Nutrition, Children’s Hospital of Philadelphia, Philadelphia, 12 PA, USA 13 14 3Department of Pathobiology, School of Veterinary Medicine, University of Pennsylvania, Philadelphia, PA, 15 USA 16 17 18 Corresponding author: Laurel Redding, [email protected] 19 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.05.11.443551; this version posted May 11, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 20 Abstract 21 Diarrheal disease, a major cause of morbidity and mortality in dairy calves, is strongly associated with the 22 health and composition of the gut microbiome. Clostridioides difficile is an opportunistic pathogen that 23 proliferates and can produce enterotoxins when the host experiences gut dysbiosis. -

Analyse Bibliographique Sur Le Microbiote Intestinal Et Son Etude Dans Des Modeles Animaux De Maladies Metaboliques, En Particulier Chez Le Primate Non Humain These
VETAGRO SUP CAMPUS VETERINAIRE DE LYON Année 2019 - Thèse n°111 ANALYSE BIBLIOGRAPHIQUE SUR LE MICROBIOTE INTESTINAL ET SON ETUDE DANS DES MODELES ANIMAUX DE MALADIES METABOLIQUES, EN PARTICULIER CHEZ LE PRIMATE NON HUMAIN THESE Présentée à l’UNIVERSITE CLAUDE-BERNARD - LYON I (Médecine - Pharmacie) et soutenue publiquement le 6 décembre 2019 pour obtenir le grade de Docteur Vétérinaire par SCHUTZ Charlotte Née le 5 mars 1994 à Zürich (Suisse) VETAGRO SUP CAMPUS VETERINAIRE DE LYON Année 2019 - Thèse n°111 ANALYSE BIBLIOGRAPHIQUE SUR LE MICROBIOTE INTESTINAL ET SON ETUDE DANS DES MODELES ANIMAUX DE MALADIES METABOLIQUES, EN PARTICULIER CHEZ LE PRIMATE NON HUMAIN THESE Présentée à l’UNIVERSITE CLAUDE-BERNARD - LYON I (Médecine - Pharmacie) et soutenue publiquement le 6 décembre 2019 pour obtenir le grade de Docteur Vétérinaire par SCHUTZ Charlotte Née le 5 mars 1994 à Zürich (Suisse) Liste du corps enseignant Liste des Enseignants du Campus Vétérinaire de Lyon (01-09-2019) ABITBOL Marie DEPT-BASIC-SCIENCES Professeur ALVES-DE-OLIVEIRA Laurent DEPT-BASIC-SCIENCES Maître de conférences ARCANGIOLI Marie-Anne DEPT-ELEVAGE-SPV Professeur AYRAL Florence DEPT-ELEVAGE-SPV Maître de conférences BECKER Claire DEPT-ELEVAGE-SPV Maître de conférences BELLUCO Sara DEPT-AC-LOISIR-SPORT Maître de conférences BENAMOU-SMITH Agnès DEPT-AC-LOISIR-SPORT Maître de conférences BENOIT Etienne DEPT-BASIC-SCIENCES Professeur BERNY Philippe DEPT-BASIC-SCIENCES Professeur BONNET-GARIN Jeanne-Marie DEPT-BASIC-SCIENCES Professeur BOULOCHER Caroline DEPT-BASIC-SCIENCES -

The Isolation of Novel Lachnospiraceae Strains and the Evaluation of Their Potential Roles in Colonization Resistance Against Clostridium Difficile
The isolation of novel Lachnospiraceae strains and the evaluation of their potential roles in colonization resistance against Clostridium difficile Diane Yuan Wang Honors Thesis in Biology Department of Ecology and Evolutionary Biology College of Literature, Science, & the Arts University of Michigan, Ann Arbor April 1st, 2014 Sponsor: Vincent B. Young, M.D., Ph.D. Associate Professor of Internal Medicine Associate Professor of Microbiology and Immunology Medical School Co-Sponsor: Aaron A. King, Ph.D. Associate Professor of Ecology & Evolutionary Associate Professor of Mathematics College of Literature, Science, & the Arts Reader: Blaise R. Boles, Ph.D. Assistant Professor of Molecular, Cellular and Developmental Biology College of Literature, Science, & the Arts 1 Table of Contents Abstract 3 Introduction 4 Clostridium difficile 4 Colonization Resistance 5 Lachnospiraceae 6 Objectives 7 Materials & Methods 9 Sample Collection 9 Bacterial Isolation and Selective Growth Conditions 9 Design of Lachnospiraceae 16S rRNA-encoding gene primers 9 DNA extraction and 16S ribosomal rRNA-encoding gene sequencing 10 Phylogenetic analyses 11 Direct inhibition 11 Bile salt hydrolase (BSH) detection 12 PCR assay for bile acid 7α-dehydroxylase detection 12 Tables & Figures Table 1 13 Table 2 15 Table 3 21 Table 4 25 Figure 1 16 Figure 2 19 Figure 3 20 Figure 4 24 Figure 5 26 Results 14 Isolation of novel Lachnospiraceae strains 14 Direct inhibition 17 Bile acid physiology 22 Discussion 27 Acknowledgments 33 References 34 2 Abstract Background: Antibiotic disruption of the gastrointestinal tract’s indigenous microbiota can lead to one of the most common nosocomial infections, Clostridium difficile, which has an annual cost exceeding $4.8 billion dollars. -

Significance of Donor Human Milk
fmicb-09-01376 June 26, 2018 Time: 17:31 # 1 ORIGINAL RESEARCH published: 27 June 2018 doi: 10.3389/fmicb.2018.01376 Preterm Gut Microbiome Depending on Feeding Type: Significance of Donor Human Milk Anna Parra-Llorca1, María Gormaz1,2, Cristina Alcántara3, María Cernada1,2, Antonio Nuñez-Ramiro1,2, Máximo Vento1,2*† and Maria C. Collado3*† 1 Neonatal Research Group, Health Research Institute La Fe, University and Polytechnic Hospital La Fe, Valencia, Spain, 2 Division of Neonatology, University and Polytechnic Hospital La Fe, Valencia, Spain, 3 Department of Biotechnology, Institute of Agrochemistry and Food Technology, Spanish National Research Council, Valencia, Spain Preterm microbial colonization is affected by gestational age, antibiotic treatment, type of birth, but also by type of feeding. Breast milk has been acknowledged as the gold standard for human nutrition. In preterm infants breast milk has been associated with improved growth and cognitive development and a reduced risk of necrotizing enterocolitis and late onset sepsis. In the absence of their mother’s own milk (MOM), pasteurized donor human milk (DHM) could be the best available alternative due to its similarity to the former. However, little is known about the effect of DHM upon preterm Edited by: Sandra Torriani, microbiota and potential biological implications. Our objective was to determine the University of Verona, Italy impact of DHM upon preterm gut microbiota admitted in a referral neonatal intensive Reviewed by: care unit (NICU). A prospective observational cohort study in NICU of 69 neonates Carlotta De Filippo, <32 weeks of gestation and with a birth weight ≤1,500 g was conducted. -
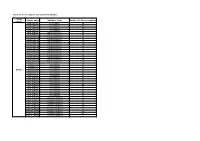
Table S1: List of Samples Included in the Analysis
Table S1: list of samples included in the analysis Study Sample name Inhibitory status Number of days at sampling number DNA.0P2T4 No inhibition 29 DNA.0P2T6 No inhibition 57 DNA.10P2T4 No inhibition 29 DNA.10P2T6 No inhibition 57 DNA.75P2T4 Phenol inhibition 29 DNA.75P2T6 Phenol inhibition 57 DNA.100P2T4 Phenol inhibition 29 DNA.100P2T6 Phenol inhibition 57 DNA.125P1T4 Phenol inhibition 29 DNA.125P1T6 Phenol inhibition 57 DNA.125P2T4 Phenol inhibition 29 DNA.125P2T6 Phenol inhibition 57 DNA.125P3T4 Phenol inhibition 29 DNA.125P3T6 Phenol inhibition 57 DNA.150P2T4 Phenol inhibition 29 DNA.150P2T6 Phenol inhibition 57 DNA.200P2T4 Phenol inhibition 29 DNA.200P2T6 Phenol inhibition 57 DNA.0N2T4 No inhibition 29 DNA.0N2T5 No inhibition 42 DNA.0N2T6 No inhibition 57 Study 1 DNA.5N2T4 No inhibition 29 DNA.5N2T5 No inhibition 42 DNA.5N2T6 No inhibition 57 DNA.10N2T4 No inhibition 29 DNA.10N2T5 No inhibition 42 DNA.10N2T6 No inhibition 57 DNA.15N2T4 No inhibition 29 DNA.15N2T5 No inhibition 42 DNA.15N2T6 No inhibition 57 DNA.25N2T4 No inhibition 29 DNA.25N2T5 No inhibition 42 DNA.25N2T6 No inhibition 57 DNA.75N2T4 Ammonia inhibition 29 DNA.75N2T5 Ammonia inhibition 42 DNA.75N2T6 Ammonia inhibition 57 DNA.100N2T4 Ammonia inhibition 29 DNA.100N2T5 Ammonia inhibition 42 DNA.100N2T6 Ammonia inhibition 57 DNA.250N2T4 Ammonia inhibition 29 DNA.250N2T5 Ammonia inhibition 42 DNA.250N2T6 Ammonia inhibition 57 nono2T3 No inhibition 16 noN2T4 Ammonia inhibition 23 noN2T8 Ammonia inhibition 60 noN2T9 Ammonia inhibition 85 noPhi2T4 Phenol inhibition 23 noPhi2T5 -

Variations of the Intestinal Gut Microbiota of Farmed Rainbow Trout, Oncorhynchus Mykiss (Walbaum), Depending on the Infection Status of the Fish"
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by IRTA Pubpro This is the peer reviewed version of the following article: Parshukov, A.N., E.N. Kashinskaya, E.P. Simonov, O.V. Hlunov, G.I. Izvekova, K.B. Andree, and M.M. Solovyev. 2019. "Variations Of The Intestinal Gut Microbiota Of Farmed Rainbow Trout, Oncorhynchus Mykiss (Walbaum), Depending On The Infection Status Of The Fish". Journal Of Applied Microbiology. Wiley. doi:10.1111/jam.14302., which has been published in final form at https://doi.org/10.1111/jam.14302. This article may be used for non-commercial purposes in accordance with Wiley Terms and Conditions for Use of Self-Archived Versions http://www.wileyauthors.com/self- archiving. DR. ALEKSEY PARSHUKOV (Orcid ID : 0000-0001-9917-186X) MISS ELENA KASHINSKAYA (Orcid ID : 0000-0001-8097-2333) Article type : - Original Article Variations of the intestinal gut microbiota of farmed rainbow trout, Oncorhynchus mykiss (Walbaum), depending on the infection status of the fish Article A.N. Parshukov1*¥, E.N. Kashinskaya2¥, E.P. Simonov2,3¥, O.V. Hlunov4, G.I. Izvekova5, K.B. Andree6, M.M. Solovyev2,7** 1Institute of Biology of the Karelian Research Centre of the Russian Academy of Sciences, Petrozavodsk, Russia 2Institute of Systematics and Ecology of Animals, Siberian Branch, Russian Academy of Sciences, Novosibirsk, Russia 3Laboratory for Genomic Research and Biotechnology, Krasnoyarsk Science Center of the Siberian Branch of the Russian Academy of Sciences, Krasnoyarsk, Russia 4LLC “FishForel”, Lahdenpohja, Karelia, Russia 5Papanin Institute for Biology of Inland Waters, Russian Academy of Sciences, Borok, Nekouzskii raion, Yaroslavl oblast, Russia 6IRTA-SCR, San Carlos de la Rapita, Tarragona, Spain 7Tomsk State University, Tomsk, Russia This article has been accepted for publication and undergone full peer review but has not Accepted been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record.