Significance of Donor Human Milk
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Fatty Acid Diets: Regulation of Gut Microbiota Composition and Obesity and Its Related Metabolic Dysbiosis
International Journal of Molecular Sciences Review Fatty Acid Diets: Regulation of Gut Microbiota Composition and Obesity and Its Related Metabolic Dysbiosis David Johane Machate 1, Priscila Silva Figueiredo 2 , Gabriela Marcelino 2 , Rita de Cássia Avellaneda Guimarães 2,*, Priscila Aiko Hiane 2 , Danielle Bogo 2, Verônica Assalin Zorgetto Pinheiro 2, Lincoln Carlos Silva de Oliveira 3 and Arnildo Pott 1 1 Graduate Program in Biotechnology and Biodiversity in the Central-West Region of Brazil, Federal University of Mato Grosso do Sul, Campo Grande 79079-900, Brazil; [email protected] (D.J.M.); [email protected] (A.P.) 2 Graduate Program in Health and Development in the Central-West Region of Brazil, Federal University of Mato Grosso do Sul, Campo Grande 79079-900, Brazil; pri.fi[email protected] (P.S.F.); [email protected] (G.M.); [email protected] (P.A.H.); [email protected] (D.B.); [email protected] (V.A.Z.P.) 3 Chemistry Institute, Federal University of Mato Grosso do Sul, Campo Grande 79079-900, Brazil; [email protected] * Correspondence: [email protected]; Tel.: +55-67-3345-7416 Received: 9 March 2020; Accepted: 27 March 2020; Published: 8 June 2020 Abstract: Long-term high-fat dietary intake plays a crucial role in the composition of gut microbiota in animal models and human subjects, which affect directly short-chain fatty acid (SCFA) production and host health. This review aims to highlight the interplay of fatty acid (FA) intake and gut microbiota composition and its interaction with hosts in health promotion and obesity prevention and its related metabolic dysbiosis. -

Effect of Ph and Temperature on Microbial Community Structure And
Calicioglu et al. Biotechnol Biofuels (2018) 11:275 https://doi.org/10.1186/s13068-018-1278-6 Biotechnology for Biofuels RESEARCH Open Access Efect of pH and temperature on microbial community structure and carboxylic acid yield during the acidogenic digestion of duckweed Ozgul Calicioglu1, Michael J. Shreve1, Tom L. Richard2 and Rachel A. Brennan1* Abstract Background: Duckweeds (Lemnaceae) are efcient aquatic plants for wastewater treatment due to their high nutri- ent-uptake capabilities and resilience to severe environmental conditions. Combined with their rapid growth rates, high starch, and low lignin contents, duckweeds have also gained popularity as a biofuel feedstock for thermochemi- cal conversion and alcohol fermentation. However, studies on the acidogenic anaerobic digestion of duckweed into carboxylic acids, another group of chemicals which are precursors of higher-value chemicals and biofuels, are lacking. In this study, a series of laboratory batch experiments were performed to determine the favorable operating condi- tions (i.e., temperature and pH) to maximize carboxylic acid production from wastewater-derived duckweed during acidogenic digestion. Batch reactors with 25 g/l solid loading were operated anaerobically for 21 days under meso- philic (35 °C) or thermophilic (55 °C) conditions at an acidic (5.3) or basic (9.2) pH. At the conclusion of the experiment, the dominant microbial communities under various operating conditions were assessed using high-throughput sequencing. Results: The highest duckweed–carboxylic acid conversion of 388 28 mg acetic acid equivalent per gram volatile solids was observed under mesophilic and basic conditions, with an± average production rate of 0.59 g/l/day. This result is comparable to those reported for acidogenic digestion of other organics such as food waste. -

Analyzing the Early Stages of Clostridium Difficile Spore
ANALYZING THE EARLY STAGES OF CLOSTRIDIUM DIFFICILE SPORE GERMINATION A Dissertation by MICHAEL FRANCIS Submitted to the Office of Graduate and Professional Studies of Texas A&M University in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Chair of Committee, Joseph A. Sorg Committee Members, James L. Smith Matthew S. Sachs Paul D. Straight Head of Department, Thomas D. McKnight May 2017 Major Subject: Microbiology Copyright 2017 Michael Francis ABSTRACT Infections caused by Clostridium difficile have increased steadily over the past several years. While studies on C. difficile virulence and physiology have been hindered, in the past, by lack of genetic approaches and suitable animal models, newly developed technologies and animal models allow for improved experimental detail. One such advance was the generation of a mouse-model of C. difficile infection. This system was an important step forward in the analysis of the genetic requirements for colonization and infection. Equally important is understanding the differences that exist between mice and humans. One of these differences is the natural bile acid composition. Bile acid-mediated spore germination, a process whereby a dormant spore returns to active, vegetative growth, is an important step during C. difficile colonization. Mice produce several different bile acids that are not found in humans (muricholic acids) that have the potential to impact C. difficile spore germination. In order to understand potential effects of these different bile acids on C. difficile physiology, we characterized their effects on C. difficile spore germination and growth of vegetative cells. We found that the mouse-derived muricholic acids affect germination similarly to a previously-described inhibitor of germination, chenodeoxycholic acid. -

Resilience of Microbial Communities After Hydrogen Peroxide Treatment of a Eutrophic Lake to Suppress Harmful Cyanobacterial Blooms
microorganisms Article Resilience of Microbial Communities after Hydrogen Peroxide Treatment of a Eutrophic Lake to Suppress Harmful Cyanobacterial Blooms Tim Piel 1,†, Giovanni Sandrini 1,†,‡, Gerard Muyzer 1 , Corina P. D. Brussaard 1,2 , Pieter C. Slot 1, Maria J. van Herk 1, Jef Huisman 1 and Petra M. Visser 1,* 1 Department of Freshwater and Marine Ecology, Institute for Biodiversity and Ecosystem Dynamics, University of Amsterdam, 1090 GE Amsterdam, The Netherlands; [email protected] (T.P.); [email protected] (G.S.); [email protected] (G.M.); [email protected] (C.P.D.B.); [email protected] (P.C.S.); [email protected] (M.J.v.H.); [email protected] (J.H.) 2 Department of Marine Microbiology and Biogeochemistry, NIOZ Royal Netherland Institute for Sea Research, 1790 AB Den Burg, The Netherlands * Correspondence: [email protected]; Tel.: +31-20-5257073 † These authors have contributed equally to this work. ‡ Current address: Department of Technology & Sources, Evides Water Company, 3006 AL Rotterdam, The Netherlands. Abstract: Applying low concentrations of hydrogen peroxide (H2O2) to lakes is an emerging method to mitigate harmful cyanobacterial blooms. While cyanobacteria are very sensitive to H2O2, little Citation: Piel, T.; Sandrini, G.; is known about the impacts of these H2O2 treatments on other members of the microbial com- Muyzer, G.; Brussaard, C.P.D.; Slot, munity. In this study, we investigated changes in microbial community composition during two P.C.; van Herk, M.J.; Huisman, J.; −1 lake treatments with low H2O2 concentrations (target: 2.5 mg L ) and in two series of controlled Visser, P.M. -

Production of Bio-Hydrogen from Dairy Wastewater Using Pretreated Landfill
Journal of Bioscience and Bioengineering VOL. 127 No. 2, 150e159, 2019 www.elsevier.com/locate/jbiosc Production of bio-hydrogen from dairy wastewater using pretreated landfill leachate sludge as an inoculum Yee Meng Wong,1,2 Pau Loke Show,3 Ta Yeong Wu,4 Hui Yi Leong,3 Shaliza Ibrahim,5 and Joon Ching Juan1,2,* School of Science, Monash University Malaysia, Jalan Lagoon Selatan, Bandar Sunway, 46150 Selangor Darul Ehsan, Malaysia,1 Nanotechnology & Catalysis Research Centre (NANOCAT), University of Malaya, 50603 Kuala Lumpur, Malaysia,2 Bioseparation Research Group, Department of Chemical and Environmental Engineering, Faculty of Engineering, University of Nottingham Malaysia Campus, Jalan Broga, 43500 Semenyih, Selangor Darul Ehsan, Malaysia,3 Chemical Engineering Discipline, School of Engineering, Monash University, Jalan Lagoon Selatan, Bandar Sunway, 46150 Selangor Darul Ehsan, Malaysia,4 and Department of Civil Engineering, Faculty of Engineering, University of Malaya, 50603 Kuala Lumpur, Malaysia5 Received 26 February 2018; accepted 15 July 2018 Available online 15 September 2018 Bio-hydrogen production from wastewater using sludge as inoculum is a sustainable approach for energy production. This study investigated the influence of initial pH and temperature on bio-hydrogen production from dairy wastewater using pretreated landfill leachate sludge (LLS) as an inoculum. The maximum yield of 113.2 ± 2.9 mmol H2/g chemical oxygen demand (COD) (12.8 ± 0.3 mmol H2/g carbohydrates) was obtained at initial pH 6 and 37 C. The main products of volatile fatty acids were acetate and butyrate with the ratio of acetate:butyrate was 0.4. At optimum condition, Gibb’s free energy was estimated at L40 kJ/mol, whereas the activation enthalpy and entropy were 65 kJ/mol and 0.128 kJ/mol/ l, respectively. -

Clostridium Amazonitimonense, Clostridium Me
ORIGINAL ARTICLE Taxonogenomic description of four new Clostridium species isolated from human gut: ‘Clostridium amazonitimonense’, ‘Clostridium merdae’, ‘Clostridium massilidielmoense’ and ‘Clostridium nigeriense’ M. T. Alou1, S. Ndongo1, L. Frégère1, N. Labas1, C. Andrieu1, M. Richez1, C. Couderc1, J.-P. Baudoin1, J. Abrahão2, S. Brah3, A. Diallo1,4, C. Sokhna1,4, N. Cassir1, B. La Scola1, F. Cadoret1 and D. Raoult1,5 1) Aix-Marseille Université, Unité de Recherche sur les Maladies Infectieuses et Tropicales Emergentes, UM63, CNRS 7278, IRD 198, INSERM 1095, Marseille, France, 2) Laboratório de Vírus, Departamento de Microbiologia, Universidade Federal de Minas Gerais, Belo Horizonte, Minas Gerais, Brazil, 3) Hopital National de Niamey, BP 247, Niamey, Niger, 4) Campus Commun UCAD-IRD of Hann, Route des pères Maristes, Hann Maristes, BP 1386, CP 18524, Dakar, Senegal and 5) Special Infectious Agents Unit, King Fahd Medical Research Center, King Abdulaziz University, Jeddah, Saudi Arabia Abstract Culturomics investigates microbial diversity of the human microbiome by combining diversified culture conditions, matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and 16S rRNA gene identification. The present study allowed identification of four putative new Clostridium sensu stricto species: ‘Clostridium amazonitimonense’ strain LF2T, ‘Clostridium massilidielmoense’ strain MT26T, ‘Clostridium nigeriense’ strain Marseille-P2414T and ‘Clostridium merdae’ strain Marseille-P2953T, which we describe using the concept of taxonogenomics. We describe the main characteristics of each bacterium and present their complete genome sequence and annotation. © 2017 Published by Elsevier Ltd. Keywords: ‘Clostridium amazonitimonense’, ‘Clostridium massilidielmoense’, ‘Clostridium merdae’, ‘Clostridium nigeriense’, culturomics, emerging bacteria, human microbiota, taxonogenomics Original Submission: 18 August 2017; Revised Submission: 9 November 2017; Accepted: 16 November 2017 Article published online: 22 November 2017 intestine [1,4–6]. -

Guide to Turning an RDP File Into a Data Set
Guide to Turning an RDP File into a Data Set Berkley Shands, Patricio S. La Rosa, Elena Deych, William Shannon July 5, 2017 Below we will define the basic steps required to generate a data set from an RDP file 1. Take an RDP file such as this example: ;Root:1.0;Bacteria:1.0;Firmicutes:1.0;Bacilli:1.0;Bacillales:1.0;Staphylococcaceae:1.0; ;Root:1.0;Bacteria:1.0;Firmicutes:1.0;Bacilli:1.0;Lactobacillales:1.0;Carnobacteriaceae:0.8; ;Root:1.0;Bacteria:1.0;Bacteroidetes:1.0;Bacteroidia:1.0;Bacteroidales:1.0;Prevotellaceae:1.0; ;Root:1.0;Bacteria:1.0;Bacteroidetes:1.0;Bacteroidia:1.0;Bacteroidales:1.0;Prevotellaceae:1.0; ;Root:1.0;Bacteria:1.0;Bacteroidetes:1.0;Bacteroidia:1.0;Unclassified:0.5;Unclassified:0.5; 2. Take the first entry and seperate each level into its own row, while seper- ating levels by a period: Root, 1 Root.Bacteria, 1 Root.Bacteria.Firmicutes, 1 Root.Bacteria.Firmicutes.Bacilli, 1 Root.Bacteria.Firmicutes.Bacilli.Bacillales, 1 Root.Bacteria.Firmicutes.Bacilli.Bacillales.Staphylococcaceae, 1 3. Do the same with each following row, adding to the number at the end if it is the same: Root, 5 Root.Bacteria, 5 Root.Bacteria.Firmicutes, 2 Root.Bacteria.Firmicutes.Bacilli, 2 Root.Bacteria.Firmicutes.Bacilli.Bacillales, 1 Root.Bacteria.Firmicutes.Bacilli.Bacillales.Staphylococcaceae, 1 Root.Bacteria.Firmicutes.Bacilli.Lactobacillales, 1 Root.Bacteria.Firmicutes.Bacilli.Lactobacillales.Carnobacteriaceae, .8 Root.Bacteria.Bacteroidetes, 3 Root.Bacteria.Bacteroidetes.Bacteroidia, 3 Root.Bacteria.Bacteroidetes.Bacteroidia.Bacteroidales, 2 Root.Bacteria.Bacteroidetes.Bacteroidia.Bacteroidales.Prevotellaceae, 2 Root.Bacteria.Bacteroidetes.Bacteroidia.Unclassified, .5 Root.Bacteria.Bacteroidetes.Bacteroidia.Unclassified.Unclassified, .5 4. -
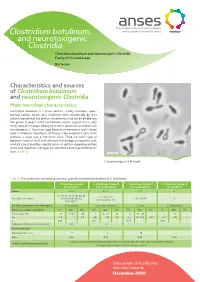
Clostridium Botulinum, and Neurotoxigenic Clostridia Clostridium Botulinum and Neurotoxigenic Clostridia Family of Clostridiaceae Bacterium
Clostridium botulinum, and neurotoxigenic Clostridia Clostridium botulinum and neurotoxigenic Clostridia Family of Clostridiaceae Bacterium Characteristics and sources of Clostridium botulinum and neurotoxigenic Clostridia Main microbial characteristics Clostridium botulinum is a Gram-positive, strictly anaerobic, spore- forming bacillus. Strains of C. botulinum differ considerably by their cultural, biochemical and genetic characteristics and can be divided into four groups (Groups I to IV). Furthermore, certain atypical strains, only rarely isolated in Europe, belonging to other species of Clostridium, are neurotoxigenic: C. butyricum (type E botulinum neurotoxin) and C. baratii (type F botulinum neurotoxin). With only a few exceptions, each strain produces a single type of botulinum toxin. There are seven types of botulinum toxin (A to G) with different immunological properties, each of which is neutralised by a specific serum. In addition, depending on their amino acid sequences, sub-types are identified in each type of botulinum toxin (Table 1). C. botulinum type A. © M. Popoff Table 1. Characteristics concerning survival, growth and toxin production of C. botulinum C. botulinum Group I C. botulinum Group II C. botulinum Group III C. botulinum Group IV (proteolytic) (non-proteolytic) (non-proteolytic) (proteolytic) Toxins A, B, F B, E, F C, D G A1, A2, A3, A4, A5, B1, B2, B3, E1, E2, E3, E6, Sous-types de toxines bivalent B (Ba, Bf, Ab), C, D, C/D, D/C G non-proteolytic B, F proteolytic F Bactéries apparentées non toxinogènes C. sporogenes Pas de nom d’espèces C. novyi C. subterminale Croissance cellules végétatives Min. Opt. Max. Min. Opt. Max. Min. Opt. Max. Min. -

Enzymatic Potential of Bacteria and Fungi Isolates from the Sewage Sludge Composting Process
applied sciences Article Enzymatic Potential of Bacteria and Fungi Isolates from the Sewage Sludge Composting Process 1,2, 1,2 1,2, Tatiana Robledo-Mahón y , Concepción Calvo and Elisabet Aranda * 1 Institute of Water Research, University of Granada, Ramón y Cajal 4, 18071 Granada, Spain; [email protected] (T.R.-M.); [email protected] (C.C.) 2 Department of Microbiology, Pharmacy Faculty, University of Granada, Campus de Cartuja s/n, 18071 Granada, Spain * Correspondence: [email protected] Current address: Department of Agro-Environmental Chemistry and Plant Nutrition, y Faculty of Agrobiology, Food and Natural Resources, Czech University of Life Sciences, Kamýcká 129, 16500 Prague 6-Suchdol, Czech Republic. Received: 6 October 2020; Accepted: 2 November 2020; Published: 3 November 2020 Featured Application: Screening of suitable microorganisms adapted to environmental conditions are the challenges for the optimization of biotechnological processes in the current and near future. Some of the isolated microorganisms in this study possess biotechnological desirable features which could be employed in different processes, such as biorefinery, bioremediation, or in the food industry. Abstract: The aim of this study was the isolation and characterisation of the fungi and bacteria during the composting process of sewage sludge under a semipermeable membrane system at full scale, in order to find isolates with enzymatic activities of biotechnological interest. A total of 40 fungi were isolated and enzymatically analysed. Fungal culture showed a predominance of members of Ascomycota and Basidiomycota division and some representatives of Mucoromycotina subdivision. Some noticeable fungi isolated during the mesophilic and thermophilic phase were Aspergillus, Circinella, and Talaromyces. -

Type of the Paper (Article
Supplementary Materials S1 Clinical details recorded, Sampling, DNA Extraction of Microbial DNA, 16S rRNA gene sequencing, Bioinformatic pipeline, Quantitative Polymerase Chain Reaction Clinical details recorded In addition to the microbial specimen, the following clinical features were also recorded for each patient: age, gender, infection type (primary or secondary, meaning initial or revision treatment), pain, tenderness to percussion, sinus tract and size of the periapical radiolucency, to determine the correlation between these features and microbial findings (Table 1). Prevalence of all clinical signs and symptoms (except periapical lesion size) were recorded on a binary scale [0 = absent, 1 = present], while the size of the radiolucency was measured in millimetres by two endodontic specialists on two- dimensional periapical radiographs (Planmeca Romexis, Coventry, UK). Sampling After anaesthesia, the tooth to be treated was isolated with a rubber dam (UnoDent, Essex, UK), and field decontamination was carried out before and after access opening, according to an established protocol, and shown to eliminate contaminating DNA (Data not shown). An access cavity was cut with a sterile bur under sterile saline irrigation (0.9% NaCl, Mölnlycke Health Care, Göteborg, Sweden), with contamination control samples taken. Root canal patency was assessed with a sterile K-file (Dentsply-Sirona, Ballaigues, Switzerland). For non-culture-based analysis, clinical samples were collected by inserting two paper points size 15 (Dentsply Sirona, USA) into the root canal. Each paper point was retained in the canal for 1 min with careful agitation, then was transferred to −80ºC storage immediately before further analysis. Cases of secondary endodontic treatment were sampled using the same protocol, with the exception that specimens were collected after removal of the coronal gutta-percha with Gates Glidden drills (Dentsply-Sirona, Switzerland). -

Updates on the Sporulation Process in Clostridium Species
Updates on the sporulation process in Clostridium species Talukdar, P. K., Olguín-Araneda, V., Alnoman, M., Paredes-Sabja, D., & Sarker, M. R. (2015). Updates on the sporulation process in Clostridium species. Research in Microbiology, 166(4), 225-235. doi:10.1016/j.resmic.2014.12.001 10.1016/j.resmic.2014.12.001 Elsevier Accepted Manuscript http://cdss.library.oregonstate.edu/sa-termsofuse *Manuscript 1 Review article for publication in special issue: Genetics of toxigenic Clostridia 2 3 Updates on the sporulation process in Clostridium species 4 5 Prabhat K. Talukdar1, 2, Valeria Olguín-Araneda3, Maryam Alnoman1, 2, Daniel Paredes-Sabja1, 3, 6 Mahfuzur R. Sarker1, 2. 7 8 1Department of Biomedical Sciences, College of Veterinary Medicine and 2Department of 9 Microbiology, College of Science, Oregon State University, Corvallis, OR. U.S.A; 3Laboratorio 10 de Mecanismos de Patogénesis Bacteriana, Departamento de Ciencias Biológicas, Facultad de 11 Ciencias Biológicas, Universidad Andrés Bello, Santiago, Chile. 12 13 14 Running Title: Clostridium spore formation. 15 16 17 Key Words: Clostridium, spores, sporulation, Spo0A, sigma factors 18 19 20 Corresponding author: Dr. Mahfuzur Sarker, Department of Biomedical Sciences, College of 21 Veterinary Medicine, Oregon State University, 216 Dryden Hall, Corvallis, OR 97331. Tel: 541- 22 737-6918; Fax: 541-737-2730; e-mail: [email protected] 23 1 24 25 Abstract 26 Sporulation is an important strategy for certain bacterial species within the phylum Firmicutes to 27 survive longer periods of time in adverse conditions. All spore-forming bacteria have two phases 28 in their life; the vegetative form, where they can maintain all metabolic activities and replicate to 29 increase numbers, and the spore form, where no metabolic activities exist. -
![Downloaded from the Ribosomal Database Project Website [22], and Aligned Using MUSCLE [23]](https://docslib.b-cdn.net/cover/9248/downloaded-from-the-ribosomal-database-project-website-22-and-aligned-using-muscle-23-2689248.webp)
Downloaded from the Ribosomal Database Project Website [22], and Aligned Using MUSCLE [23]
Diversity 2013, 5, 627-640; doi:10.3390/d5030627 OPEN ACCESS diversity ISSN 1424-2818 www.mdpi.com/journal/diversity Article Untangling the Genetic Basis of Fibrolytic Specialization by Lachnospiraceae and Ruminococcaceae in Diverse Gut Communities Amy Biddle 1, Lucy Stewart 1, Jeffrey Blanchard 2 and Susan Leschine 3,* 1 Department of Microbiology, University of Massachusetts, Amherst, MA 01003, USA; E-Mails: [email protected] (A.B.); [email protected] (L.S.) 2 Department of Biology, University of Massachusetts, Amherst, MA 01003, USA; E-Mail: [email protected] 3 Department of Veterinary and Animal Sciences, University of Massachusetts, Amherst, MA 01003, USA * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +1-413-545-0673; Fax: +1-413-545-6326. Received: 17 May 2013; in revised form: 21 June 2013 / Accepted: 26 June 2013 / Published: 9 August 2013 Abstract: The Lachnospiraceae and Ruminococcaceae are two of the most abundant families from the order Clostridiales found in the mammalian gut environment, and have been associated with the maintenance of gut health. While they are both diverse groups, they share a common role as active plant degraders. By comparing the genomes of the Lachnospiraceae and Ruminococcaceae with the Clostridiaceae, a more commonly free-living group, we identify key carbohydrate-active enzymes, sugar transport mechanisms, and metabolic pathways that distinguish these two commensal groups as specialists for the degradation of complex plant material. Keywords: Clostridiales; Ruminococcaceae; Lachnospiraceae; carbohydrate-active enzymes; comparative genomics; plant degradation Diversity 2013, 5 628 1. Introduction 1.1. Taxonomic Revision of the Clostridiales Is a Work in Progress Classically, the genus Clostridium was described as comprising spore-forming, non-sulfate reducing obligate anaerobic bacteria with a gram-positive cell wall.