Heat Resistant Thermophilic Endospores in Cold Estuarine Sediments
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

High-Quality Draft Genome Sequences of Five Anaerobic Oral Bacteria and Description of Peptoanaerobacter Stomatis Gen
Sizova et al. Standards in Genomic Sciences (2015) 10:37 DOI 10.1186/s40793-015-0027-8 EXTENDED GENOME REPORT Open Access High-quality draft genome sequences of five anaerobic oral bacteria and description of Peptoanaerobacter stomatis gen. nov., sp. nov., a new member of the family Peptostreptococcaceae Maria V. Sizova1, Amanda Chilaka1, Ashlee M. Earl2, Sebastian N. Doerfert1, Paul A. Muller1, Manolito Torralba3, Jamison M. McCorrison3, A. Scott Durkin3, Karen E. Nelson3 and Slava S. Epstein1* Abstract Here we report a summary classification and the features of five anaerobic oral bacteria from the family Peptostreptococcaceae. Bacterial strains were isolated from human subgingival plaque. Strains ACC19a, CM2, CM5, and OBRC8 represent the first known cultivable members of “yet uncultured” human oral taxon 081; strain AS15 belongs to “cultivable” human oral taxon 377. Based on 16S rRNA gene sequence comparisons, strains ACC19a, CM2, CM5, and OBRC8 are distantly related to Eubacterium yurii subs. yurii and Filifactor alocis, with 93.2 – 94.4 % and 85.5 % of sequence identity, respectively. The genomes of strains ACC19a, CM2, CM5, OBRC8 and AS15 are 2,541,543; 2,312,592; 2,594,242; 2,553,276; and 2,654,638 bp long. The genomes are comprised of 2277, 1973, 2325, 2277, and 2308 protein-coding genes and 54, 57, 54, 36, and 28 RNA genes, respectively. Based on the distinct characteristics presented here, we suggest that strains ACC19a, CM2, CM5, and OBRC8 represent a novel genus and species within the family Peptostreptococcaceae, for which we propose the name Peptoanaerobacter stomatis gen. nov., sp. nov. The type strain is strain ACC19aT (=HM-483T; =DSM 28705T;=ATCC BAA-2665T). -

Microbial Community Structure Dynamics in Ohio River Sediments During Reductive Dechlorination of Pcbs
University of Kentucky UKnowledge University of Kentucky Doctoral Dissertations Graduate School 2008 MICROBIAL COMMUNITY STRUCTURE DYNAMICS IN OHIO RIVER SEDIMENTS DURING REDUCTIVE DECHLORINATION OF PCBS Andres Enrique Nunez University of Kentucky Right click to open a feedback form in a new tab to let us know how this document benefits ou.y Recommended Citation Nunez, Andres Enrique, "MICROBIAL COMMUNITY STRUCTURE DYNAMICS IN OHIO RIVER SEDIMENTS DURING REDUCTIVE DECHLORINATION OF PCBS" (2008). University of Kentucky Doctoral Dissertations. 679. https://uknowledge.uky.edu/gradschool_diss/679 This Dissertation is brought to you for free and open access by the Graduate School at UKnowledge. It has been accepted for inclusion in University of Kentucky Doctoral Dissertations by an authorized administrator of UKnowledge. For more information, please contact [email protected]. ABSTRACT OF DISSERTATION Andres Enrique Nunez The Graduate School University of Kentucky 2008 MICROBIAL COMMUNITY STRUCTURE DYNAMICS IN OHIO RIVER SEDIMENTS DURING REDUCTIVE DECHLORINATION OF PCBS ABSTRACT OF DISSERTATION A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the College of Agriculture at the University of Kentucky By Andres Enrique Nunez Director: Dr. Elisa M. D’Angelo Lexington, KY 2008 Copyright © Andres Enrique Nunez 2008 ABSTRACT OF DISSERTATION MICROBIAL COMMUNITY STRUCTURE DYNAMICS IN OHIO RIVER SEDIMENTS DURING REDUCTIVE DECHLORINATION OF PCBS The entire stretch of the Ohio River is under fish consumption advisories due to contamination with polychlorinated biphenyls (PCBs). In this study, natural attenuation and biostimulation of PCBs and microbial communities responsible for PCB transformations were investigated in Ohio River sediments. Natural attenuation of PCBs was negligible in sediments, which was likely attributed to low temperature conditions during most of the year, as well as low amounts of available nitrogen, phosphorus, and organic carbon. -

Genome-Resolved Meta-Analysis of the Microbiome in Oil Reservoirs Worldwide
microorganisms Article Genome-Resolved Meta-Analysis of the Microbiome in Oil Reservoirs Worldwide Kelly J. Hidalgo 1,2,* , Isabel N. Sierra-Garcia 3 , German Zafra 4 and Valéria M. de Oliveira 1 1 Microbial Resources Division, Research Center for Chemistry, Biology and Agriculture (CPQBA), University of Campinas–UNICAMP, Av. Alexandre Cazellato 999, 13148-218 Paulínia, Brazil; [email protected] 2 Graduate Program in Genetics and Molecular Biology, Institute of Biology, University of Campinas (UNICAMP), Rua Monteiro Lobato 255, Cidade Universitária, 13083-862 Campinas, Brazil 3 Biology Department & CESAM, University of Aveiro, Aveiro, Portugal, Campus de Santiago, Avenida João Jacinto de Magalhães, 3810-193 Aveiro, Portugal; [email protected] 4 Grupo de Investigación en Bioquímica y Microbiología (GIBIM), Escuela de Microbiología, Universidad Industrial de Santander, Cra 27 calle 9, 680002 Bucaramanga, Colombia; [email protected] * Correspondence: [email protected]; Tel.: +55-19981721510 Abstract: Microorganisms inhabiting subsurface petroleum reservoirs are key players in biochemical transformations. The interactions of microbial communities in these environments are highly complex and still poorly understood. This work aimed to assess publicly available metagenomes from oil reservoirs and implement a robust pipeline of genome-resolved metagenomics to decipher metabolic and taxonomic profiles of petroleum reservoirs worldwide. Analysis of 301.2 Gb of metagenomic information derived from heavily flooded petroleum reservoirs in China and Alaska to non-flooded petroleum reservoirs in Brazil enabled us to reconstruct 148 metagenome-assembled genomes (MAGs) of high and medium quality. At the phylum level, 74% of MAGs belonged to bacteria and 26% to archaea. The profiles of these MAGs were related to the physicochemical parameters and recovery management applied. -

Supplementary Information
doi: 10.1038/nature06269 SUPPLEMENTARY INFORMATION METAGENOMIC AND FUNCTIONAL ANALYSIS OF HINDGUT MICROBIOTA OF A WOOD FEEDING HIGHER TERMITE TABLE OF CONTENTS MATERIALS AND METHODS 2 • Glycoside hydrolase catalytic domains and carbohydrate binding modules used in searches that are not represented by Pfam HMMs 5 SUPPLEMENTARY TABLES • Table S1. Non-parametric diversity estimators 8 • Table S2. Estimates of gross community structure based on sequence composition binning, and conserved single copy gene phylogenies 8 • Table S3. Summary of numbers glycosyl hydrolases (GHs) and carbon-binding modules (CBMs) discovered in the P3 luminal microbiota 9 • Table S4. Summary of glycosyl hydrolases, their binning information, and activity screening results 13 • Table S5. Comparison of abundance of glycosyl hydrolases in different single organism genomes and metagenome datasets 17 • Table S6. Comparison of abundance of glycosyl hydrolases in different single organism genomes (continued) 20 • Table S7. Phylogenetic characterization of the termite gut metagenome sequence dataset, based on compositional phylogenetic analysis 23 • Table S8. Counts of genes classified to COGs corresponding to different hydrogenase families 24 • Table S9. Fe-only hydrogenases (COG4624, large subunit, C-terminal domain) identified in the P3 luminal microbiota. 25 • Table S10. Gene clusters overrepresented in termite P3 luminal microbiota versus soil, ocean and human gut metagenome datasets. 29 • Table S11. Operational taxonomic unit (OTU) representatives of 16S rRNA sequences obtained from the P3 luminal fluid of Nasutitermes spp. 30 SUPPLEMENTARY FIGURES • Fig. S1. Phylogenetic identification of termite host species 38 • Fig. S2. Accumulation curves of 16S rRNA genes obtained from the P3 luminal microbiota 39 • Fig. S3. Phylogenetic diversity of P3 luminal microbiota within the phylum Spirocheates 40 • Fig. -

WO 2018/064165 A2 (.Pdf)
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2018/064165 A2 05 April 2018 (05.04.2018) W !P O PCT (51) International Patent Classification: Published: A61K 35/74 (20 15.0 1) C12N 1/21 (2006 .01) — without international search report and to be republished (21) International Application Number: upon receipt of that report (Rule 48.2(g)) PCT/US2017/053717 — with sequence listing part of description (Rule 5.2(a)) (22) International Filing Date: 27 September 2017 (27.09.2017) (25) Filing Language: English (26) Publication Langi English (30) Priority Data: 62/400,372 27 September 2016 (27.09.2016) US 62/508,885 19 May 2017 (19.05.2017) US 62/557,566 12 September 2017 (12.09.2017) US (71) Applicant: BOARD OF REGENTS, THE UNIVERSI¬ TY OF TEXAS SYSTEM [US/US]; 210 West 7th St., Austin, TX 78701 (US). (72) Inventors: WARGO, Jennifer; 1814 Bissonnet St., Hous ton, TX 77005 (US). GOPALAKRISHNAN, Vanch- eswaran; 7900 Cambridge, Apt. 10-lb, Houston, TX 77054 (US). (74) Agent: BYRD, Marshall, P.; Parker Highlander PLLC, 1120 S. Capital Of Texas Highway, Bldg. One, Suite 200, Austin, TX 78746 (US). (81) Designated States (unless otherwise indicated, for every kind of national protection available): AE, AG, AL, AM, AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, HR, HU, ID, IL, IN, IR, IS, JO, JP, KE, KG, KH, KN, KP, KR, KW, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. -

ATP Hydrolysis by a Domain Related to Translation Factor Gtpases Drives
ATP hydrolysis by a domain related to translation PNAS PLUS factor GTPases drives polymerization of a static bacterial morphogenetic protein Jean-Philippe Castainga, Attila Nagyb,1, Vivek Anantharamanc,1, L. Aravindc, and Kumaran S. Ramamurthia,2 aLaboratory of Molecular Biology, National Cancer Institute, National Institutes of Health, Bethesda, MD 20892; bLaboratory of Molecular Physiology, National Heart, Lung and Blood Institute, National Institutes of Health, Bethesda, MD 20892; and cNational Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20894 Edited by E. Peter Greenberg, University of Washington, Seattle, WA, and approved November 28, 2012 (received for review June 21, 2012) The assembly of static supramolecular structures is a culminating the coat assembles (20) and whose structural component is event of developmental programs. One such structure, the pro- composed of a protein called SpoIVA (pronounced “Spo-four-A”; teinaceous shell (called the coat) that surrounds spores of the hereafter called IVA) (16, 21, 22) (Fig. 1A). IVA is anchored bacterium Bacillus subtilis, is composed of about 70 different pro- to the surface of the forespore by a small amphipathic protein teins and represents one of the most durable biological structures that dictates the correct subcellular location of IVA (22–26), and known. The coat is built atop a basement layer that contains an its encasement around the forespore depends on a soluble protein ATPase (SpoIVA) that forms a platform required for coat assembly. in the mother cell (27). Previously, we reported that IVA binds Here, we show that SpoIVA belongs to the translation factors class and hydrolyzes ATP in vitro and that disruption of a “Walker A” of P-loop GTPases and has evolutionarily lost the ability to bind motif in IVA, required for ATP binding, disrupted sporulation GTP; instead, it uses ATP hydrolysis to drive its self-assembly into efficiency of cells producing the variant protein in vivo (28). -

Horizontal Gene Flow Into Geobacillus Is Constrained by the Chromosomal Organization of Growth and Sporulation
bioRxiv preprint doi: https://doi.org/10.1101/381442; this version posted August 2, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Horizontal gene flow into Geobacillus is constrained by the chromosomal organization of growth and sporulation Alexander Esin1,2, Tom Ellis3,4, Tobias Warnecke1,2* 1Molecular Systems Group, Medical Research Council London Institute of Medical Sciences, London, United Kingdom 2Institute of Clinical Sciences, Faculty of Medicine, Imperial College London, London, United Kingdom 3Imperial College Centre for Synthetic Biology, Imperial College London, London, United Kingdom 4Department of Bioengineering, Imperial College London, London, United Kingdom *corresponding author ([email protected]) 1 bioRxiv preprint doi: https://doi.org/10.1101/381442; this version posted August 2, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Abstract Horizontal gene transfer (HGT) in bacteria occurs in the context of adaptive genome architecture. As a consequence, some chromosomal neighbourhoods are likely more permissive to HGT than others. Here, we investigate the chromosomal topology of horizontal gene flow into a clade of Bacillaceae that includes Geobacillus spp. Reconstructing HGT patterns using a phylogenetic approach coupled to model-based reconciliation, we discover three large contiguous chromosomal zones of HGT enrichment. -
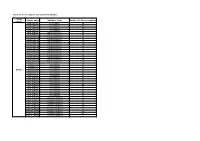
Table S1: List of Samples Included in the Analysis
Table S1: list of samples included in the analysis Study Sample name Inhibitory status Number of days at sampling number DNA.0P2T4 No inhibition 29 DNA.0P2T6 No inhibition 57 DNA.10P2T4 No inhibition 29 DNA.10P2T6 No inhibition 57 DNA.75P2T4 Phenol inhibition 29 DNA.75P2T6 Phenol inhibition 57 DNA.100P2T4 Phenol inhibition 29 DNA.100P2T6 Phenol inhibition 57 DNA.125P1T4 Phenol inhibition 29 DNA.125P1T6 Phenol inhibition 57 DNA.125P2T4 Phenol inhibition 29 DNA.125P2T6 Phenol inhibition 57 DNA.125P3T4 Phenol inhibition 29 DNA.125P3T6 Phenol inhibition 57 DNA.150P2T4 Phenol inhibition 29 DNA.150P2T6 Phenol inhibition 57 DNA.200P2T4 Phenol inhibition 29 DNA.200P2T6 Phenol inhibition 57 DNA.0N2T4 No inhibition 29 DNA.0N2T5 No inhibition 42 DNA.0N2T6 No inhibition 57 Study 1 DNA.5N2T4 No inhibition 29 DNA.5N2T5 No inhibition 42 DNA.5N2T6 No inhibition 57 DNA.10N2T4 No inhibition 29 DNA.10N2T5 No inhibition 42 DNA.10N2T6 No inhibition 57 DNA.15N2T4 No inhibition 29 DNA.15N2T5 No inhibition 42 DNA.15N2T6 No inhibition 57 DNA.25N2T4 No inhibition 29 DNA.25N2T5 No inhibition 42 DNA.25N2T6 No inhibition 57 DNA.75N2T4 Ammonia inhibition 29 DNA.75N2T5 Ammonia inhibition 42 DNA.75N2T6 Ammonia inhibition 57 DNA.100N2T4 Ammonia inhibition 29 DNA.100N2T5 Ammonia inhibition 42 DNA.100N2T6 Ammonia inhibition 57 DNA.250N2T4 Ammonia inhibition 29 DNA.250N2T5 Ammonia inhibition 42 DNA.250N2T6 Ammonia inhibition 57 nono2T3 No inhibition 16 noN2T4 Ammonia inhibition 23 noN2T8 Ammonia inhibition 60 noN2T9 Ammonia inhibition 85 noPhi2T4 Phenol inhibition 23 noPhi2T5 -

Microbial Diversity of Drilling Fluids from 3000 M Deep Koyna Pilot
Science Reports Sci. Dril., 27, 1–23, 2020 https://doi.org/10.5194/sd-27-1-2020 © Author(s) 2020. This work is distributed under the Creative Commons Attribution 4.0 License. Microbial diversity of drilling fluids from 3000 m deep Koyna pilot borehole provides insights into the deep biosphere of continental earth crust Himadri Bose1, Avishek Dutta1, Ajoy Roy2, Abhishek Gupta1, Sourav Mukhopadhyay1, Balaram Mohapatra1, Jayeeta Sarkar1, Sukanta Roy3, Sufia K. Kazy2, and Pinaki Sar1 1Environmental Microbiology and Genomics Laboratory, Department of Biotechnology, Indian Institute of Technology Kharagpur, Kharagpur, 721302, West Bengal, India 2Department of Biotechnology, National Institute of Technology Durgapur, Durgapur, 713209, West Bengal, India 3Ministry of Earth Sciences, Borehole Geophysics Research Laboratory, Karad, 415114, Maharashtra, India Correspondence: Pinaki Sar ([email protected], [email protected]) Received: 24 June 2019 – Revised: 25 September 2019 – Accepted: 16 December 2019 – Published: 27 May 2020 Abstract. Scientific deep drilling of the Koyna pilot borehole into the continental crust up to a depth of 3000 m below the surface at the Deccan Traps, India, provided a unique opportunity to explore microbial life within the deep granitic bedrock of the Archaean Eon. Microbial communities of the returned drilling fluid (fluid re- turned to the mud tank from the underground during the drilling operation; designated here as DF) sampled during the drilling operation of the Koyna pilot borehole at a depth range of 1681–2908 metres below the surface (m b.s.) were explored to gain a glimpse of the deep biosphere underneath the continental crust. Change of pH 2− to alkalinity, reduced abundance of Si and Al, but enrichment of Fe, Ca and SO4 in the samples from deeper horizons suggested a gradual infusion of elements or ions from the crystalline bedrock, leading to an observed geochemical shift in the DF. -

Food Processing and Preservation - Sbt1607
SCHOOL OF BIO AND CHEMICAL ENGINEERING DEPARTMENT OF BIOTECHNOLOGY B.TECH – BIOTECHNOLOGY UNIT – I - FOOD PROCESSING AND PRESERVATION - SBT1607 HISTORY OF FOOD PROCESSING AND FOOD PRESERVATION FOOD PROCESSING Food processing dates back to the prehistoric age when crude processing including various types of cooking, such as over fire, smoking, steaming, fermenting, sun drying and preserving with salt were in practice. Foods preserved this way were a common part of warriors’ and sailors’ diets. These crude processing techniques remained essentially the same until the advent of the Industrial Revolution. Nicolas Appert developed a vacuum bottling process to supply food to troops in the French army, which eventually led to canning in tins by Peter Durand in 1810. Modern food processing technologies, in the 19th century were also largely developed to serve military needs. In the early 20th century, the space race, change in food habits and the quality conciousness of the consumers in the developed world furthered the development of food processing with advancements such as spray drying, juice concentrates, freeze drying and the introduction of artificial sweetners, colourants, and preservatives. In the late 20th century products including dried instant soups, reconstituted fruit juices, and self cooking meals such as ready-to-eat food rations etc., were developed. Benefits of Processing . Converts raw food and other farm produce into edible, usable and palatable form. Helps to store perishable and semi-perishable agricultural commodities, avoid glut in the market, check post harvest losses and make the produce available during off-season. Generates employment. Development of ready-to-consume products, hence saves time for cooking. -

Genome Analysis of Desulfotomaculum Gibsoniae Strain Grollt a Highly Versatile Gram-Positive Sulfate-Reducing Bacterium
UvA-DARE (Digital Academic Repository) Genome analysis of Desulfotomaculum gibsoniae strain GrollT a highly versatile Gram-positive sulfate-reducing bacterium Kuever, J.; Visser, M.; Loeffler, C.; Boll, M.; Worm, P.; Sousa, D.Z.; Plugge, C.M.; Schaap, P.J.; Muyzer, G.; Pereira, I.A.C.; Parshina, S.N.; Goodwin, L.A; Kyrpides, N.C.; Detter, J.; Woyke, T.; Chain, P.; Davenport, K.W.; Rohde, M.; Spring, S.; Klenk, H.-P.; Stams, A.J.M. DOI 10.4056/sigs.5209235 Publication date 2014 Document Version Final published version Published in Standards in Genomic Sciences Link to publication Citation for published version (APA): Kuever, J., Visser, M., Loeffler, C., Boll, M., Worm, P., Sousa, D. Z., Plugge, C. M., Schaap, P. J., Muyzer, G., Pereira, I. A. C., Parshina, S. N., Goodwin, L. A., Kyrpides, N. C., Detter, J., Woyke, T., Chain, P., Davenport, K. W., Rohde, M., Spring, S., ... Stams, A. J. M. (2014). Genome analysis of Desulfotomaculum gibsoniae strain GrollT a highly versatile Gram- positive sulfate-reducing bacterium. Standards in Genomic Sciences, 9(3), 821-839. https://doi.org/10.4056/sigs.5209235 General rights It is not permitted to download or to forward/distribute the text or part of it without the consent of the author(s) and/or copyright holder(s), other than for strictly personal, individual use, unless the work is under an open content license (like Creative Commons). Disclaimer/Complaints regulations If you believe that digital publication of certain material infringes any of your rights or (privacy) interests, please let the Library know, stating your reasons. -

Complete Genome Sequence of the Hyperthermophilic Bacteria- Thermotoga Sp
COMPLETE GENOME SEQUENCE OF THE HYPERTHERMOPHILIC BACTERIA- THERMOTOGA SP. STRAIN RQ7 Rutika Puranik A Thesis Submitted to the Graduate College of Bowling Green State University in partial fulfillment of the requirements for the degree of MASTER OF SCIENCE May 2015 Committee: Zhaohui Xu, Advisor Scott Rogers George Bullerjahn © 2015 Rutika Puranik All Rights Reserved iii ABSTRACT Zhaohui Xu, Advisor The genus Thermotoga is one of the deep-rooted genus in the phylogenetic tree of life and has been studied for its thermostable enzymes and the property of hydrogen production at higher temperatures. The current study focuses on the complete genome sequencing of T. sp. strain RQ7 to understand and identify the conserved as well as variable properties between the strains and its genus with the approach of comparative genomics. A pipeline was developed to assemble the complete genome based on the next generation sequencing (NGS) data. The pipeline successfully combined computational approaches with wet lab experiments to deliver a completed genome of T. sp. strain RQ7 that has the genome size of 1,851,618 bp with a GC content of 47.1%. The genome is submitted to Genbank with accession CP07633. Comparative genomic analysis of this genome with three other strains of Thermotoga, helped identifying putative natural transformation and competence protein coding genes in addition to the absence of TneDI restriction- modification system in T. sp. strain RQ7. Genome analysis also assisted in recognizing the unique genes in T. sp. strain RQ7 and CRISPR/Cas system. This strain has 8 CRISPR loci and an array of Cas coding genes in the entire genome.