Horizontal Gene Flow Into Geobacillus Is Constrained by the Chromosomal Organization of Growth and Sporulation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Transcriptional Landscape of a Rewritten Bacterial Genome Reveals Control Elements and Genome Design Principles ✉ ✉ Mariëlle J
ARTICLE https://doi.org/10.1038/s41467-021-23362-y OPEN The transcriptional landscape of a rewritten bacterial genome reveals control elements and genome design principles ✉ ✉ Mariëlle J. F. M. van Kooten 1 , Clio A. Scheidegger1, Matthias Christen1 & Beat Christen 1 Sequence rewriting enables low-cost genome synthesis and the design of biological systems with orthogonal genetic codes. The error-free, robust rewriting of nucleotide sequences can 1234567890():,; be achieved with a complete annotation of gene regulatory elements. Here, we compare transcription in Caulobacter crescentus to transcription from plasmid-borne segments of the synthesized genome of C. ethensis 2.0. This rewritten derivative contains an extensive amount of supposedly neutral mutations, including 123’562 synonymous codon changes. The tran- scriptional landscape refines 60 promoter annotations, exposes 18 termination elements and links extensive transcription throughout the synthesized genome to the unintentional intro- duction of sigma factor binding motifs. We reveal translational regulation for 20 CDS and uncover an essential translational regulatory element for the expression of ribosomal protein RplS. The annotation of gene regulatory elements allowed us to formulate design principles that improve design schemes for synthesized DNA, en route to a bright future of iteration- free programming of biological systems. 1 Institute of Molecular Systems Biology, Department of Biology, Eidgenössische Technische Hochschule Zürich, Zürich, Switzerland. ✉ email: [email protected]; [email protected] NATURE COMMUNICATIONS | (2021) 12:3053 | https://doi.org/10.1038/s41467-021-23362-y | www.nature.com/naturecommunications 1 ARTICLE NATURE COMMUNICATIONS | https://doi.org/10.1038/s41467-021-23362-y e can program biological systems with DNA that is In previous work, we synthesized and assembled the rewritten Wbased on native nucleotide sequences. -

Appendix III: OTU's Found to Be Differentially Abundant Between CD and Control Patients Via Metagenomeseq Analysis
Appendix III: OTU's found to be differentially abundant between CD and control patients via metagenomeSeq analysis OTU Log2 (FC CD / FDR Adjusted Phylum Class Order Family Genus Species Number Control) p value 518372 Firmicutes Clostridia Clostridiales Ruminococcaceae Faecalibacterium prausnitzii 2.16 5.69E-08 194497 Firmicutes Clostridia Clostridiales Ruminococcaceae NA NA 2.15 8.93E-06 175761 Firmicutes Clostridia Clostridiales Ruminococcaceae NA NA 5.74 1.99E-05 193709 Firmicutes Clostridia Clostridiales Ruminococcaceae NA NA 2.40 2.14E-05 4464079 Bacteroidetes Bacteroidia Bacteroidales Bacteroidaceae Bacteroides NA 7.79 0.000123188 20421 Firmicutes Clostridia Clostridiales Lachnospiraceae Coprococcus NA 1.19 0.00013719 3044876 Firmicutes Clostridia Clostridiales Lachnospiraceae [Ruminococcus] gnavus -4.32 0.000194983 184000 Firmicutes Clostridia Clostridiales Ruminococcaceae Faecalibacterium prausnitzii 2.81 0.000306032 4392484 Bacteroidetes Bacteroidia Bacteroidales Bacteroidaceae Bacteroides NA 5.53 0.000339948 528715 Firmicutes Clostridia Clostridiales Ruminococcaceae Faecalibacterium prausnitzii 2.17 0.000722263 186707 Firmicutes Clostridia Clostridiales NA NA NA 2.28 0.001028539 193101 Firmicutes Clostridia Clostridiales Ruminococcaceae NA NA 1.90 0.001230738 339685 Firmicutes Clostridia Clostridiales Peptococcaceae Peptococcus NA 3.52 0.001382447 101237 Firmicutes Clostridia Clostridiales NA NA NA 2.64 0.001415109 347690 Firmicutes Clostridia Clostridiales Ruminococcaceae Oscillospira NA 3.18 0.00152075 2110315 Firmicutes Clostridia -

Heat Resistant Thermophilic Endospores in Cold Estuarine Sediments
Heat resistant thermophilic endospores in cold estuarine sediments Emma Bell Thesis submitted for the degree of Doctor of Philosophy School of Civil Engineering and Geosciences Faculty of Science, Agriculture and Engineering February 2016 Abstract Microbial biogeography explores the spatial and temporal distribution of microorganisms at multiple scales and is influenced by environmental selection and passive dispersal. Understanding the relative contribution of these factors can be challenging as their effects can be difficult to differentiate. Dormant thermophilic endospores in cold sediments offer a natural model for studies focusing on passive dispersal. Understanding distributions of these endospores is not confounded by the influence of environmental selection; rather their occurrence is due exclusively to passive transport. Sediment heating experiments were designed to investigate the dispersal histories of various thermophilic spore-forming Firmicutes in the River Tyne, a tidal estuary in North East England linking inland tributaries with the North Sea. Microcosm incubations at 50-80°C were monitored for sulfate reduction and enriched bacterial populations were characterised using denaturing gradient gel electrophoresis, functional gene clone libraries and high-throughput sequencing. The distribution of thermophilic endospores among different locations along the estuary was spatially variable, indicating that dispersal vectors originating in both warm terrestrial and marine habitats contribute to microbial diversity in estuarine and marine environments. In addition to their persistence in cold sediments, some endospores displayed a remarkable heat-resistance surviving multiple rounds of autoclaving. These extremely heat-resistant endospores are genetically similar to those detected in deep subsurface environments, including geothermal groundwater investigated from a nearby terrestrial borehole drilled to >1800 m depth with bottom temperatures in excess of 70°C. -

Distribution and Characteristics of Bacillus Bacteria Associated with Hydrobionts and the Waters of the Peter the Great Bay, Sea of Japan I
ISSN 0026-2617, Microbiology, 2008, Vol. 77, No. 4, pp. 497–503. © Pleiades Publishing, Ltd., 2008. Original Russian Text © I.A. Beleneva, 2008, published in Mikrobiologiya, 2008, Vol. 77, No. 4, pp. 558–565. EXPERIMENTAL ARTICLES Distribution and Characteristics of Bacillus Bacteria Associated with Hydrobionts and the Waters of the Peter the Great Bay, Sea of Japan I. A. Beleneva1 Zhirmunskii Institute of Marine Biology, Far East Division, Russian Academy of Sciences, ul. Pal’chevskogo, 17, Vladivostok 690041, Russia Received May 28, 2007 Abstract—Bacilli of the species Bacillus subtilis, B. pumilus, B. mycoides, B. marinus and B. licheniformis (a total of 53 strains) were isolated from 15 invertebrate species and the water of the Vostok Bay, Peter the Great Bay, Sea of Japan. Bacilli were most often isolated from bivalves (22.7%) and sea cucumbers (18.9%); they occurred less frequently in sea urchins and starfish (13.2 and 7.5%, respectively). Most of bacilli strains were isolated from invertebrates inhabiting silted sediments. No Bacillus spp. strains were isolated from invertebrates inhabiting stony and sandy environments. The species diversity of bacilli isolated from marine objects under study was low. Almost all bacterial isolates were resistant to lincomycin. Unlike B. pumilus, B. subtilis isolates were mostly resistant to benzylpenicillin and ampicillin. Antibiotic sensitivity of B. licheniformis strains was variable (two strains were resistant to benzylpenicillin and oxacillin, while one was sensitive). A significant fraction of isolated bacilli contained pigments. Pigmented strains were more often isolated from seawater sam- ples, while colorless ones predominated within hydrobionts. B. subtilis colonies had the broadest range of co- lors. -

Probiotic Lactic Acid Bacteria Vs. Bacilli: Pros and Cons ARTHUR C
Probiotic lactic acid bacteria vs. bacilli: pros and cons ARTHUR C. OUWEHAND1*, SOFIA FORSSTEN1, MARKUS LEHTINEN1, ELIZABETH GALBRAITH2, ELLEN DAVIS2 *Corresponding author 1. Active Nutrition, DuPont Nutrition & Health, Sokeritehtaantie 20, 02460 Kantvik, Finland Arthur C. Ouwehand 2. Animal and Environmental Applications, DuPont Nutrition & Health, W227 N752 Westmound Drive, Waukesha, WI 53186, USA KEYWORDS: probiotic, lactic acid bacteria, Lactobacillus, Bifidobacterium, Bacillus, spores ABSTRACT: Both lactic acid bacteria (LAB) and bacilli are commercialised as probiotics; LAB are mainly utilised as live and active or dormant cells, while bacilli are utilised as spores. Belonging to very different genera, they have different technological properties. LAB have a longer history of use as probiotics and therefore have a broader base as far as health efficacy documentation is concerned. The question therefore sometimes arises; strains from which of these groups make better probiotics? Considering the fact that probiotics, by definition, have to be viable in sufficient numbers and have to have documented health benefits, this is not a relevant question. Any microbial strain has to fulfil these criteria in order to qualify as a probiotic. The current paper reviews and compares the available data for both groups of organisms. Pre - Probiotics INTRODUCTION In order to supply consumers with products that provide meaningful levels of probiotics, several requirements have There is no legal description of probiotics. However, the most to be fulfilled by probiotic strains selected for industrial commonly accepted definition is from a FAO/WHO working production. Quality-control must be vigilantly performed, group: “Live microorganisms which when administered in assuring identity, viability and absence of contaminants. -

Section 4. Guidance Document on Horizontal Gene Transfer Between Bacteria
306 - PART 2. DOCUMENTS ON MICRO-ORGANISMS Section 4. Guidance document on horizontal gene transfer between bacteria 1. Introduction Horizontal gene transfer (HGT) 1 refers to the stable transfer of genetic material from one organism to another without reproduction. The significance of horizontal gene transfer was first recognised when evidence was found for ‘infectious heredity’ of multiple antibiotic resistance to pathogens (Watanabe, 1963). The assumed importance of HGT has changed several times (Doolittle et al., 2003) but there is general agreement now that HGT is a major, if not the dominant, force in bacterial evolution. Massive gene exchanges in completely sequenced genomes were discovered by deviant composition, anomalous phylogenetic distribution, great similarity of genes from distantly related species, and incongruent phylogenetic trees (Ochman et al., 2000; Koonin et al., 2001; Jain et al., 2002; Doolittle et al., 2003; Kurland et al., 2003; Philippe and Douady, 2003). There is also much evidence now for HGT by mobile genetic elements (MGEs) being an ongoing process that plays a primary role in the ecological adaptation of prokaryotes. Well documented is the example of the dissemination of antibiotic resistance genes by HGT that allowed bacterial populations to rapidly adapt to a strong selective pressure by agronomically and medically used antibiotics (Tschäpe, 1994; Witte, 1998; Mazel and Davies, 1999). MGEs shape bacterial genomes, promote intra-species variability and distribute genes between distantly related bacterial genera. Horizontal gene transfer (HGT) between bacteria is driven by three major processes: transformation (the uptake of free DNA), transduction (gene transfer mediated by bacteriophages) and conjugation (gene transfer by means of plasmids or conjugative and integrated elements). -

Multi-Product Lactic Acid Bacteria Fermentations: a Review
fermentation Review Multi-Product Lactic Acid Bacteria Fermentations: A Review José Aníbal Mora-Villalobos 1 ,Jéssica Montero-Zamora 1, Natalia Barboza 2,3, Carolina Rojas-Garbanzo 3, Jessie Usaga 3, Mauricio Redondo-Solano 4, Linda Schroedter 5, Agata Olszewska-Widdrat 5 and José Pablo López-Gómez 5,* 1 National Center for Biotechnological Innovations of Costa Rica (CENIBiot), National Center of High Technology (CeNAT), San Jose 1174-1200, Costa Rica; [email protected] (J.A.M.-V.); [email protected] (J.M.-Z.) 2 Food Technology Department, University of Costa Rica (UCR), San Jose 11501-2060, Costa Rica; [email protected] 3 National Center for Food Science and Technology (CITA), University of Costa Rica (UCR), San Jose 11501-2060, Costa Rica; [email protected] (C.R.-G.); [email protected] (J.U.) 4 Research Center in Tropical Diseases (CIET) and Food Microbiology Section, Microbiology Faculty, University of Costa Rica (UCR), San Jose 11501-2060, Costa Rica; [email protected] 5 Bioengineering Department, Leibniz Institute for Agricultural Engineering and Bioeconomy (ATB), 14469 Potsdam, Germany; [email protected] (L.S.); [email protected] (A.O.-W.) * Correspondence: [email protected]; Tel.: +49-(0331)-5699-857 Received: 15 December 2019; Accepted: 4 February 2020; Published: 10 February 2020 Abstract: Industrial biotechnology is a continuously expanding field focused on the application of microorganisms to produce chemicals using renewable sources as substrates. Currently, an increasing interest in new versatile processes, able to utilize a variety of substrates to obtain diverse products, can be observed. -
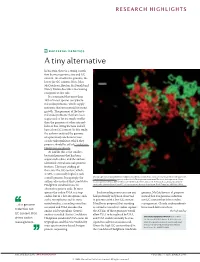
Bacterial Genetics a Tiny Alternative
RESEARCH HIGHLIGHTS BACTERIAL GENETICS A tiny alternative In bacteria, there is a strong correla- tion between genome size and GC content: the smaller the genome, the lower the GC content. Now, John McCutcheon, Bradon McDonald and Nancy Moran describe a fascinating exception to this rule. It is estimated that more than 10% of insect species carry bacte- rial endosymbionts, which supply nutrients that are essential for insect growth. The genomes of the bacte- rial endosymbionts that have been sequenced so far are much smaller than the genomes of other intracel- lular or free-living bacteria and all have a low GC content. In this study, the authors analysed the genome of a previously uncharacterized cicada endosymbiont, which they propose should be called Candidatus Hodgkinia cicadicola. At 144 kb, this is the smallest bacterial genome that has been sequenced to date, and the authors identified several unusual genomic features. The most striking of these was the GC content, which, at 58%, is unusually high for such Micrograph showing Candidatus Hodgkinia cicadicola (red) in close association with another endosymbiont, a small genome. Intriguingly, the Candidatus Sulcia muelleri (green) in the cicada Diceroprocta semicincta. The scale bar represents 10 µm. authors also noticed that Candidatus Image reproduced from McCutcheon, J. P., McDonald, B. R. & Moran, N. P. Origin of an alternative genetic Hodgkinia cicadicola uses an code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 5, e1000565 (2009). alternative genetic code. In most species the codon UGA is a stop Such recoding events are rare and genome, McCutcheon et al. -

Synthetic Biology Projects in Vitro
Downloaded from genome.cshlp.org on September 25, 2021 - Published by Cold Spring Harbor Laboratory Press Review Synthetic biology projects in vitro Anthony C. Forster1,3 and George M. Church2,3 1Department of Pharmacology and Vanderbilt Institute of Chemical Biology, Vanderbilt University Medical Center, Nashville, Tennessee 37232, USA; 2Department of Genetics, Harvard Medical School, Boston, Massachusetts 02115, USA Advances in the in vitro synthesis and evolution of DNA, RNA, and polypeptides are accelerating the construction of biopolymers, pathways, and organisms with novel functions. Known functions are being integrated and debugged with the aim of synthesizing life-like systems. The goals are knowledge, tools, smart materials, and therapies. Synthetic biology projects (SBPs) an action plan to regulate world suppliers of DNA synthesizers, DNA precursors, and oligos (Church 2004). Ethical and safety The basic elements of chemistry and biology are few, but the issues have been, and must continue to be, regulated (Cho et al. synthetic combinations are unlimited and awe inspiring. The 1999). first international conference on synthetic biology charted its Several groups have proposed to create bacteria with chro- goals as understanding and utilizing life’s diverse solutions to mosomes synthesized entirely from synthetic oligos. This might process information, materials, and energy (Silver and Way 2004) be done stepwise (Posfai et al. 2006) or by inactivating the en- (http://syntheticbiology.org). As a bonus, genetic systems are dogenous bacterial chromosome and then somehow transform- biocompatible, renewable, and can be optimized by Darwinian ing and rebooting the bacterium with an entire in vitro- selections. SBPs entail the complex manipulation of replicating synthesized genome. -

Genomic and Transcriptome Analyses of a Thermophilic Bacterium
Genomic and transcriptome analyses of a thermophilic bacterium Geobacillus stearothermophilus B5 isolated from compost reveal its enzymatic basis for lignocellulose degradation Mengmeng Wang Nanjing Agricultural University Jiaxi Miao Nanjing Agricultural University Xuanqing Wang Nanjing Agricultural University Tuo Li Nanjing Agricultural University Han Zhu Nanjing Agricultural University Dongyang Liu ( [email protected] ) Nanjing Agricultural University https://orcid.org/0000-0001-7452-9423 Qirong Shen Nanjing Agricultural University Research Keywords: Geobacillus stearothermophilus, thermophilic composting, cellulose-degrading, comparative genomics, transcriptome Posted Date: March 24th, 2020 DOI: https://doi.org/10.21203/rs.3.rs-18489/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Version of Record: A version of this preprint was published at Microorganisms on September 4th, 2020. See the published version at https://doi.org/10.3390/microorganisms8091357. Page 1/23 Abstract Background Composting is a special global carbon cycle which sustains various microbes engendering the cellulose degradation. Studies have obtained substantial compost microbiomes, yet the expressions and functions of these lignocellulolytic enzymes remains obscure. Thus, the discovery of thermophilic microorganisms as considerable biochemical catalysts for biofuels is becoming more and more attractive. Results A lignocellulose degrading strain isolated from thermophilic compost was identied as Geobacillus stearothermophilus B5, which could secrete considerable enzymes at the optimal temperature (60°C) and pH (7.5). One single contig of 3.37 Mbp was obtained from raw data and 3371 protein-coding genes were predicted, and the clusters of orthologous groups (COG) analysis revealed various genes with function of polymeric substrates degradation, especially for abundant CAZymes including glycoside hydrolases (GH, 29%) and glycosyl transferases (GT, 36%). -

Nor Hawani Salikin
Characterisation of a novel antinematode agent produced by the marine epiphytic bacterium Pseudoalteromonas tunicata and its impact on Caenorhabditis elegans Nor Hawani Salikin A thesis in fulfilment of the requirements for the degree of Doctor of Philosophy School of Biological, Earth and Environmental Sciences Faculty of Science August 2020 Thesis/Dissertation Sheet Surname/Family Name : Salikin Given Name/s : Nor Hawani Abbreviation for degree as give in the University : Ph.D. calendar Faculty : UNSW Faculty of Science School : School of Biological, Earth and Environmental Sciences Characterisation of a novel antinematode agent produced Thesis Title : by the marine epiphytic bacterium Pseudoalteromonas tunicata and its impact on Caenorhabditis elegans Abstract 350 words maximum: (PLEASE TYPE) Drug resistance among parasitic nematodes has resulted in an urgent need for the development of new therapies. However, the high re-discovery rate of antinematode compounds from terrestrial environments necessitates a new repository for future drug research. Marine epiphytic bacteria are hypothesised to produce nematicidal compounds as a defence against bacterivorous predators, thus representing a promising, yet underexplored source for antinematode drug discovery. The marine epiphytic bacterium Pseudoalteromonas tunicata is known to produce a number of bioactive compounds. Screening genomic libraries of P. tunicata against the nematode Caenorhabditis elegans identified a clone (HG8) showing fast-killing activity. However, the molecular, chemical and biological properties of HG8 remain undetermined. A novel Nematode killing protein-1 (Nkp-1) encoded by an uncharacterised gene of HG8 annotated as hp1 was successfully discovered through this project. The Nkp-1 toxicity appears to be nematode-specific, with the protein being highly toxic to nematode larvae but having no impact on nematode eggs. -

Phylogenetic Study of Thermophilic Genera Anoxybacillus, Geobacillus
bioRxiv preprint doi: https://doi.org/10.1101/2021.06.18.449068; this version posted June 19, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 1 bioRxiv 2 Article Type: Research article. 3 Article Title: Phylogenetic study of thermophilic genera Anoxybacillus, Geobacillus, 4 Parageobacillus, and proposal of a new classification Quasigeobacillus 5 gen. nov. 6 Authors: Talamantes-Becerra, Berenice1-2*; Carling, Jason2; Blom, Jochen3; 7 Georges, Arthur1 8 Affiliation: 1Institute for Applied Ecology, University of Canberra ACT 2601 9 Australia. 10 2Diversity Arrays Technology Pty Ltd, Bruce, Canberra ACT 2617 11 Australia. 12 3Bioinformatics & Systems Biology, Justus-Liebig-University Giessen, 13 35392 Glessen, Hesse, Germany 14 *Correspondence: Berenice Talamantes Becerra, Institute for Applied Ecology, 15 University of Canberra ACT 2601 Australia. Email: 16 [email protected] 17 Running head: Phylogenetic study of thermophilic bacteria. 18 Word Count: Abstract: 178 Main body: 3709 References: n= 60 19 20 21 bioRxiv preprint doi: https://doi.org/10.1101/2021.06.18.449068; this version posted June 19, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. 22 23 Abstract 24 A phylogenetic study of Anoxybacillus, Geobacillus and Parageobacillus was performed using 25 publicly available whole genome sequences. A total of 113 genomes were selected for 26 phylogenomic metrics including calculation of Average Nucleotide Identity (ANI) and 27 Average Amino acid Identity (AAI), and a maximum likelihood tree was built from alignment 28 of a set of 662 orthologous core genes.