Synthetic Biology Projects in Vitro
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Transcriptional Landscape of a Rewritten Bacterial Genome Reveals Control Elements and Genome Design Principles ✉ ✉ Mariëlle J
ARTICLE https://doi.org/10.1038/s41467-021-23362-y OPEN The transcriptional landscape of a rewritten bacterial genome reveals control elements and genome design principles ✉ ✉ Mariëlle J. F. M. van Kooten 1 , Clio A. Scheidegger1, Matthias Christen1 & Beat Christen 1 Sequence rewriting enables low-cost genome synthesis and the design of biological systems with orthogonal genetic codes. The error-free, robust rewriting of nucleotide sequences can 1234567890():,; be achieved with a complete annotation of gene regulatory elements. Here, we compare transcription in Caulobacter crescentus to transcription from plasmid-borne segments of the synthesized genome of C. ethensis 2.0. This rewritten derivative contains an extensive amount of supposedly neutral mutations, including 123’562 synonymous codon changes. The tran- scriptional landscape refines 60 promoter annotations, exposes 18 termination elements and links extensive transcription throughout the synthesized genome to the unintentional intro- duction of sigma factor binding motifs. We reveal translational regulation for 20 CDS and uncover an essential translational regulatory element for the expression of ribosomal protein RplS. The annotation of gene regulatory elements allowed us to formulate design principles that improve design schemes for synthesized DNA, en route to a bright future of iteration- free programming of biological systems. 1 Institute of Molecular Systems Biology, Department of Biology, Eidgenössische Technische Hochschule Zürich, Zürich, Switzerland. ✉ email: [email protected]; [email protected] NATURE COMMUNICATIONS | (2021) 12:3053 | https://doi.org/10.1038/s41467-021-23362-y | www.nature.com/naturecommunications 1 ARTICLE NATURE COMMUNICATIONS | https://doi.org/10.1038/s41467-021-23362-y e can program biological systems with DNA that is In previous work, we synthesized and assembled the rewritten Wbased on native nucleotide sequences. -

Horizontal Gene Flow Into Geobacillus Is Constrained by the Chromosomal Organization of Growth and Sporulation
bioRxiv preprint doi: https://doi.org/10.1101/381442; this version posted August 2, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Horizontal gene flow into Geobacillus is constrained by the chromosomal organization of growth and sporulation Alexander Esin1,2, Tom Ellis3,4, Tobias Warnecke1,2* 1Molecular Systems Group, Medical Research Council London Institute of Medical Sciences, London, United Kingdom 2Institute of Clinical Sciences, Faculty of Medicine, Imperial College London, London, United Kingdom 3Imperial College Centre for Synthetic Biology, Imperial College London, London, United Kingdom 4Department of Bioengineering, Imperial College London, London, United Kingdom *corresponding author ([email protected]) 1 bioRxiv preprint doi: https://doi.org/10.1101/381442; this version posted August 2, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Abstract Horizontal gene transfer (HGT) in bacteria occurs in the context of adaptive genome architecture. As a consequence, some chromosomal neighbourhoods are likely more permissive to HGT than others. Here, we investigate the chromosomal topology of horizontal gene flow into a clade of Bacillaceae that includes Geobacillus spp. Reconstructing HGT patterns using a phylogenetic approach coupled to model-based reconciliation, we discover three large contiguous chromosomal zones of HGT enrichment. -

Section 4. Guidance Document on Horizontal Gene Transfer Between Bacteria
306 - PART 2. DOCUMENTS ON MICRO-ORGANISMS Section 4. Guidance document on horizontal gene transfer between bacteria 1. Introduction Horizontal gene transfer (HGT) 1 refers to the stable transfer of genetic material from one organism to another without reproduction. The significance of horizontal gene transfer was first recognised when evidence was found for ‘infectious heredity’ of multiple antibiotic resistance to pathogens (Watanabe, 1963). The assumed importance of HGT has changed several times (Doolittle et al., 2003) but there is general agreement now that HGT is a major, if not the dominant, force in bacterial evolution. Massive gene exchanges in completely sequenced genomes were discovered by deviant composition, anomalous phylogenetic distribution, great similarity of genes from distantly related species, and incongruent phylogenetic trees (Ochman et al., 2000; Koonin et al., 2001; Jain et al., 2002; Doolittle et al., 2003; Kurland et al., 2003; Philippe and Douady, 2003). There is also much evidence now for HGT by mobile genetic elements (MGEs) being an ongoing process that plays a primary role in the ecological adaptation of prokaryotes. Well documented is the example of the dissemination of antibiotic resistance genes by HGT that allowed bacterial populations to rapidly adapt to a strong selective pressure by agronomically and medically used antibiotics (Tschäpe, 1994; Witte, 1998; Mazel and Davies, 1999). MGEs shape bacterial genomes, promote intra-species variability and distribute genes between distantly related bacterial genera. Horizontal gene transfer (HGT) between bacteria is driven by three major processes: transformation (the uptake of free DNA), transduction (gene transfer mediated by bacteriophages) and conjugation (gene transfer by means of plasmids or conjugative and integrated elements). -
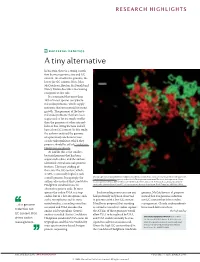
Bacterial Genetics a Tiny Alternative
RESEARCH HIGHLIGHTS BACTERIAL GENETICS A tiny alternative In bacteria, there is a strong correla- tion between genome size and GC content: the smaller the genome, the lower the GC content. Now, John McCutcheon, Bradon McDonald and Nancy Moran describe a fascinating exception to this rule. It is estimated that more than 10% of insect species carry bacte- rial endosymbionts, which supply nutrients that are essential for insect growth. The genomes of the bacte- rial endosymbionts that have been sequenced so far are much smaller than the genomes of other intracel- lular or free-living bacteria and all have a low GC content. In this study, the authors analysed the genome of a previously uncharacterized cicada endosymbiont, which they propose should be called Candidatus Hodgkinia cicadicola. At 144 kb, this is the smallest bacterial genome that has been sequenced to date, and the authors identified several unusual genomic features. The most striking of these was the GC content, which, at 58%, is unusually high for such Micrograph showing Candidatus Hodgkinia cicadicola (red) in close association with another endosymbiont, a small genome. Intriguingly, the Candidatus Sulcia muelleri (green) in the cicada Diceroprocta semicincta. The scale bar represents 10 µm. authors also noticed that Candidatus Image reproduced from McCutcheon, J. P., McDonald, B. R. & Moran, N. P. Origin of an alternative genetic Hodgkinia cicadicola uses an code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 5, e1000565 (2009). alternative genetic code. In most species the codon UGA is a stop Such recoding events are rare and genome, McCutcheon et al. -

Assessment of the Bimodality in the Distribution of Bacterial Genome Sizes
The ISME Journal (2017) 11, 821–824 © 2017 International Society for Microbial Ecology All rights reserved 1751-7362/17 www.nature.com/ismej SHORT COMMUNICATION Assessment of the bimodality in the distribution of bacterial genome sizes Hyun S Gweon, Mark J Bailey and Daniel S Read Centre for Ecology & Hydrology, Wallingford, UK Bacterial genome sizes have previously been shown to exhibit a bimodal distribution. This phenomenon has prompted discussion regarding the evolutionary forces driving genome size in bacteria and its ecological significance. We investigated the level of inherent redundancy in the public database and the effect it has on the shape of the apparent bimodal distribution. Our study reveals that there is a significant bias in the genome sequencing efforts towards a certain group of species, and that correcting the bias using species nomenclature and clustering of the 16S rRNA gene, results in a unimodal rather than the previously published bimodal distribution. The true genome size distribution and its wider ecological implications will soon emerge as we are currently witnessing rapid growth in the number of sequenced genomes from diverse environmental niches across a range of habitats at an unprecedented rate. The ISME Journal (2017) 11, 821–824; doi:10.1038/ismej.2016.142; published online 11 November 2016 Significant progress has been made in understanding Wolf in 2008, where it was reported that bacterial interactions between ecology and genome evolution genome sizes show a bimodal distribution (Koonin in prokaryotes. -

Mechanisms Of, and Barriers To, Horizontal Gene Transfer Between Bacteria
FOCUS ON HORIZONTAL GENE TRANSFER MECHANISMS OF, AND BARRIERS TO, HORIZONTAL GENE TRANSFER BETWEEN BACTERIA Christopher M. Thomas* and Kaare M. Nielsen‡ Abstract | Bacteria evolve rapidly not only by mutation and rapid multiplication, but also by transfer of DNA, which can result in strains with beneficial mutations from more than one parent. Transformation involves the release of naked DNA followed by uptake and recombination. Homologous recombination and DNA-repair processes normally limit this to DNA from similar bacteria. However, if a gene moves onto a broad-host-range plasmid it might be able to spread without the need for recombination. There are barriers to both these processes but they reduce, rather than prevent, gene acquisition. The first evidence that horizontal gene transfer (HGT) informational genes of the central cellular machinery could occur was the recognition that virulence deter- such as DNA replication, transcription or translation minants could be transferred between pneumococci in tend not to spread rapidly, even if they confer anti biotic infected mice, a phenomenon that was later shown to resistance, compared for example to single-function- be mediated by the uptake of the genetic material DNA resistance determinants such as β-lactamases or in a process called transformation1. The subsequent aminoglycoside-modifying enzymes. However, the identification of gene transfer mediated by both plas- nature of the transfer mechanism can also determine the mids and viruses and the recognition of transposable organisms and genes that are most often involved. The elements provided the stepping stones to our current purpose of this review is to describe some of the mecha- picture of gene flux and the importance of mobile nisms that lead to horizontal gene acquisitions with a genetic elements2. -

Selection and Robustness in Bacterial Genome Evolution Seila Omer University of Connecticut, [email protected]
University of Connecticut OpenCommons@UConn Doctoral Dissertations University of Connecticut Graduate School 12-16-2016 Selection and Robustness in Bacterial Genome Evolution Seila Omer University Of Connecticut, [email protected] Follow this and additional works at: https://opencommons.uconn.edu/dissertations Recommended Citation Omer, Seila, "Selection and Robustness in Bacterial Genome Evolution" (2016). Doctoral Dissertations. 1317. https://opencommons.uconn.edu/dissertations/1317 Selection and Robustness in Bacterial Genome Evolution Seila Omer, Ph.D. University of Connecticut, 2016 The research presented in this thesis attempts to address research questions related to the role of natural selection in the evolution of bacterial genes not expressed for function and in building mutational tolerance to translational errors. Studies on evolution of protein coding DNA sequences have provided the evidence for a current paradigm in evolutionary biology: only functional genes are undergoing selection against the deleterious effects of allele variants (purifying selection). I provide evidence that similar footprints of selection can be detected in genes that are not normally expressed for function during the bacterial life cycle. Using simulations for DNA sequence evolution, I demonstrate statistically significant deviations from neutral evolution for the studied genes. I suggest that purifying selection affects both functional and non-functional genes. I propose this might be caused by the dominant toxic effects of low level translation of mutated products in bacteria, due to misfolding and misinteraction. Natural selection also acts to remove the effects of translational errors. Stop codon readthrough events are more likely to have major structural and functional effects than simple nucleotide changes. Recent research has shown that strength of selection experienced by protein-coding genes is positively correlated with the level of gene expression. -

A Highly Precise and Portable Genome Engineering Method Allows Comparison of Mutational Effects Across Bacterial Species
A highly precise and portable genome engineering method allows comparison of mutational effects across bacterial species Ákos Nyergesa,1, Bálint Csörgo}a,1,2, István Nagyb,c, Balázs Bálintc, Péter Biharic, Viktória Lázára, Gábor Apjoka, Kinga Umenhoffera, Balázs Bogosa,3, György Pósfaia, and Csaba Pála,2 aSynthetic and Systems Biology Unit, Institute of Biochemistry, Biological Research Centre of the Hungarian Academy of Sciences, Szeged H-6726, Hungary; bSymbiosis and Functional Genomics Unit, Institute of Biochemistry, Biological Research Centre of the Hungarian Academy of Sciences, Szeged H-6726, Hungary; and cSeqOmics Biotechnology Ltd., Mórahalom H-6782, Hungary Edited by Roy Curtiss III, University of Florida, Gainesville, FL, and approved January 26, 2016 (received for review October 9, 2015) Currently available tools for multiplex bacterial genome engineer- rapidly creates desired allele combinations and combinatorial ge- ing are optimized for a few laboratory model strains, demand ex- nomic libraries. From accelerated optimization of biosynthetic tensive prior modification of the host strain, and lead to the pathways (5, 7–9) to the construction of a so-called “genomically accumulation of numerous off-target modifications. Building on prior recoded organism” (10–12), MAGE has allowed genome-engi- development of multiplex automated genome engineering (MAGE), neering endeavors of unparalleled complexity in Escherichia coli. our work addresses these problems in a single framework. Using a Functionality of ssDNA-mediated recombineering has been de- dominant-negative mutant protein of the methyl-directed mismatch scribed in various other species, including lactic acid bacteria (13), repair (MMR) system, we achieved a transient suppression of DNA Corynebacterium glutamicum (14), and Bacillus subtilis (15). -

Single-Pass Classification of All Noncoding Sequences in a Bacterial Genome Using Phylogenetic Profiles
Downloaded from genome.cshlp.org on September 26, 2021 - Published by Cold Spring Harbor Laboratory Press Methods Single-pass classification of all noncoding sequences in a bacterial genome using phylogenetic profiles Antonin Marchais, Magali Naville, Chantal Bohn, Philippe Bouloc, and Daniel Gautheret1 Universite´ Paris-Sud 11, CNRS, UMR8621, Institut de Ge´ne´tique et Microbiologie, F-91405 Orsay Cedex, France Identification and characterization of functional elements in the noncoding regions of genomes is an elusive and time- consuming activity whose output does not keep up with the pace of genome sequencing. Hundreds of bacterial genomes lay unexploited in terms of noncoding sequence analysis, although they may conceal a wide diversity of novel RNA genes, riboswitches, or other regulatory elements. We describe a strategy that exploits the entirety of available bacterial genomes to classify all noncoding elements of a selected reference species in a single pass. This method clusters noncoding elements based on their profile of presence among species. Most noncoding RNAs (ncRNAs) display specific signatures that enable their grouping in distinct clusters, away from sequence conservation noise and other elements such as promoters. We submitted 24 ncRNA candidates from Staphylococcus aureus to experimental validation and confirmed the presence of seven novel small RNAs or riboswitches. Besides offering a powerful method for de novo ncRNA identification, the analysis of phylogenetic profiles opens a new path toward the identification of functional relationships between co-evolving coding and noncoding elements. [Supplemental material is available online at www.genome.org.] In all living organisms, the genome regions located between served sequence detection and the presence of adjacent Rho- protein-coding sequences are home to a wide diversity of func- independent terminators. -

Chemical Synthesis Rewriting of a Bacterial Genome to Achieve Design flexibility and Biological Functionality
Chemical synthesis rewriting of a bacterial genome to achieve design flexibility and biological functionality Jonathan E. Venetza, Luca Del Medicoa, Alexander Wolfle¨ a, Philipp Schachle¨ a, Yves Buchera, Donat Apperta, Flavia Tschana, Carlos E. Flores-Tinocoa, Marielle¨ van Kootena, Rym Guennouna, Samuel Deutschb, Matthias Christena,1, and Beat Christena,1 aInstitute of Molecular Systems Biology, Eidgenossische¨ Technische Hochschule Zurich,¨ CH-8093 Zurich,¨ Switzerland; and bDepartment of Energy Joint Genome Institute, Walnut Creek, CA 94598 Edited by David Baker, University of Washington, Seattle, WA, and approved March 6, 2019 (received for review October 29, 2018) Understanding how to program biological functions into artificial been covered. The redesigned chromosomes removed repetitive DNA sequences remains a key challenge in synthetic genomics. sequences (tRNA genes, introns, and transposons) to increase Here, we report the chemical synthesis and testing of Caulobacter targeting fidelity during stepwise homologous replacement as ethensis-2.0 (C. eth-2.0), a rewritten bacterial genome composed well as included the seeding of loxP sites to permit iterative of the most fundamental functions of a bacterial cell. We rebuilt genome reduction on completion of yeast chromosomes. In the the essential genome of Caulobacter crescentus through the pro- beginning of the yeast 2.0 synthesis project, CRISPR had not yet cess of chemical synthesis rewriting and studied the genetic entered the stage, but today, it offers an alternative approach for information content at the level of its essential genes. Within progressive genome reduction. the 785,701-bp genome, we used sequence rewriting to reduce The redundancy of the genetic code defining the same amino the number of encoded genetic features from 6,290 to 799. -

Large Scale and Significant Expression from Pseudogenes in Sodalis Glossinidius - a Facultative Bacterial Endosymbiont
bioRxiv preprint doi: https://doi.org/10.1101/124388; this version posted July 21, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Large scale and significant expression from pseudogenes in Sodalis glossinidius - a facultative bacterial endosymbiont Ian Goodhead1,2, Frances Blow1,3, Philip Brownridge1 , Margaret Hughes1,4, John Kenny1,4, Ritesh Krishna1,5, Lynn MacLean1, Pisut Pongchaikul1, Rob Beynon1 and Alistair C. Darby1,4 1. Institute of Integrative Biology, University of Liverpool, Crown Street, Liverpool. L69 7ZB. UK. 2. School of Environment and Life Sciences, Peel Building, University of Salford. M5 4WT. UK. 3. Department of Entomology, Cornell University, Ithaca 14853, NY, USA. 4. Centre for Genomic Research, Institute of Integrative Biology, University of Liverpool, Crown Street, Liverpool. L69 7ZB. UK. 5. IBM Research UK, STFC Daresbury Laboratory, Warrington, WA4 4AD, UK. Abstract The majority of bacterial genomes have high coding efficiencies, but there are some genomes of intracellular bacteria that have low gene density. The genome of the endosymbiont Sodalis glossinidius contains almost 50% pseudogenes containing mutations that putatively silence them at the genomic level. We have applied multiple omic strategies, combining: Illumina and Pacific Biosciences Single-Molecule Real Time DNA-sequencing and annotation; stranded RNA-sequencing; and proteome analysis to better understand the transcriptional and translational landscape of Sodalis pseudogenes, and potential mechanisms for their control. Between 53% and 74% of the Sodalis transcriptome remains active in cell-free culture. -

Single-Pass Classification of All Non-Coding Sequences in a Bacterial Genome Using Phylogenetic Profiles
Downloaded from genome.cshlp.org on October 4, 2021 - Published by Cold Spring Harbor Laboratory Press SINGLE-PASS CLASSIFICATION OF ALL NON-CODING SEQUENCES IN A BACTERIAL GENOME USING PHYLOGENETIC PROFILES ANTONIN MARCHAIS, MAGALI NAVILLE, CHANTAL BOHN, PHILIPPE BOULOC AND DANIEL GAUTHERET1 Univ. Paris-Sud 11, CNRS, UMR8621, Institut de Génétique et Microbiologie Bâtiment 400, F-91405 Orsay Cedex, France January 25, 2009 1 Corresponding author Running title: Phylogenetic profiling of non-coding elements Keywords: non-coding RNA, ncRNA, phylogenetic profiling, small RNA, sRNA Downloaded from genome.cshlp.org on October 4, 2021 - Published by Cold Spring Harbor Laboratory Press ABSTRACT Identification and characterization of functional elements in the non-coding regions of genomes is an elusive and time consuming activity whose output does not keep up with the pace of genome sequencing. Hundreds of bacterial genomes lay unexploited in terms of non-coding sequence analysis although they may conceal a wide diversity of novel RNA genes, riboswitches or other regulatory elements. We describe a strategy that exploits the entirety of available bacterial genomes to classify all non-coding elements of a selected reference species in a single pass. This method clusters non-coding elements based on their profile of presence among species. Most non-coding RNAs display specific signatures that enable their grouping in distinct clusters, away from sequence conservation noise and other elements such as promoters. We submitted 24 ncRNA candidates from Staphylococcus aureus to experimental validation and confirmed the presence of seven novel small RNAs or riboswitches. Besides offering a powerful method for de novo ncRNA identification, the analysis of phylogenetic profiles opens a new path towards the identification of functional relationships between co-evolving coding and non-coding elements.