Chemical Synthesis Rewriting of a Bacterial Genome to Achieve Design
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

1 Evidence for Gliadin Antibodies As Causative Agents in Schizophrenia
1 Evidence for gliadin antibodies as causative agents in schizophrenia. C.J.Carter PolygenicPathways, 20 Upper Maze Hill, Saint-Leonard’s on Sea, East Sussex, TN37 0LG [email protected] Tel: 0044 (0)1424 422201 I have no fax Abstract Antibodies to gliadin, a component of gluten, have frequently been reported in schizophrenia patients, and in some cases remission has been noted following the instigation of a gluten free diet. Gliadin is a highly immunogenic protein, and B cell epitopes along its entire immunogenic length are homologous to the products of numerous proteins relevant to schizophrenia (p = 0.012 to 3e-25). These include members of the DISC1 interactome, of glutamate, dopamine and neuregulin signalling networks, and of pathways involved in plasticity, dendritic growth or myelination. Antibodies to gliadin are likely to cross react with these key proteins, as has already been observed with synapsin 1 and calreticulin. Gliadin may thus be a causative agent in schizophrenia, under certain genetic and immunological conditions, producing its effects via antibody mediated knockdown of multiple proteins relevant to the disease process. Because of such homology, an autoimmune response may be sustained by the human antigens that resemble gliadin itself, a scenario supported by many reports of immune activation both in the brain and in lymphocytes in schizophrenia. Gluten free diets and removal of such antibodies may be of therapeutic benefit in certain cases of schizophrenia. 2 Introduction A number of studies from China, Norway, and the USA have reported the presence of gliadin antibodies in schizophrenia 1-5. Gliadin is a component of gluten, intolerance to which is implicated in coeliac disease 6. -

Downloaded from the National Database for Autism Research (NDAR)
International Journal of Molecular Sciences Article Phenotypic Subtyping and Re-Analysis of Existing Methylation Data from Autistic Probands in Simplex Families Reveal ASD Subtype-Associated Differentially Methylated Genes and Biological Functions Elizabeth C. Lee y and Valerie W. Hu * Department of Biochemistry and Molecular Medicine, The George Washington University, School of Medicine and Health Sciences, Washington, DC 20037, USA; [email protected] * Correspondence: [email protected]; Tel.: +1-202-994-8431 Current address: W. Harry Feinstone Department of Molecular Microbiology and Immunology, y Johns Hopkins Bloomberg School of Public Health, Baltimore, MD 21205, USA. Received: 25 August 2020; Accepted: 17 September 2020; Published: 19 September 2020 Abstract: Autism spectrum disorder (ASD) describes a group of neurodevelopmental disorders with core deficits in social communication and manifestation of restricted, repetitive, and stereotyped behaviors. Despite the core symptomatology, ASD is extremely heterogeneous with respect to the severity of symptoms and behaviors. This heterogeneity presents an inherent challenge to all large-scale genome-wide omics analyses. In the present study, we address this heterogeneity by stratifying ASD probands from simplex families according to the severity of behavioral scores on the Autism Diagnostic Interview-Revised diagnostic instrument, followed by re-analysis of existing DNA methylation data from individuals in three ASD subphenotypes in comparison to that of their respective unaffected siblings. We demonstrate that subphenotyping of cases enables the identification of over 1.6 times the number of statistically significant differentially methylated regions (DMR) and DMR-associated genes (DAGs) between cases and controls, compared to that identified when all cases are combined. Our analyses also reveal ASD-related neurological functions and comorbidities that are enriched among DAGs in each phenotypic subgroup but not in the combined case group. -

The Transcriptional Landscape of a Rewritten Bacterial Genome Reveals Control Elements and Genome Design Principles ✉ ✉ Mariëlle J
ARTICLE https://doi.org/10.1038/s41467-021-23362-y OPEN The transcriptional landscape of a rewritten bacterial genome reveals control elements and genome design principles ✉ ✉ Mariëlle J. F. M. van Kooten 1 , Clio A. Scheidegger1, Matthias Christen1 & Beat Christen 1 Sequence rewriting enables low-cost genome synthesis and the design of biological systems with orthogonal genetic codes. The error-free, robust rewriting of nucleotide sequences can 1234567890():,; be achieved with a complete annotation of gene regulatory elements. Here, we compare transcription in Caulobacter crescentus to transcription from plasmid-borne segments of the synthesized genome of C. ethensis 2.0. This rewritten derivative contains an extensive amount of supposedly neutral mutations, including 123’562 synonymous codon changes. The tran- scriptional landscape refines 60 promoter annotations, exposes 18 termination elements and links extensive transcription throughout the synthesized genome to the unintentional intro- duction of sigma factor binding motifs. We reveal translational regulation for 20 CDS and uncover an essential translational regulatory element for the expression of ribosomal protein RplS. The annotation of gene regulatory elements allowed us to formulate design principles that improve design schemes for synthesized DNA, en route to a bright future of iteration- free programming of biological systems. 1 Institute of Molecular Systems Biology, Department of Biology, Eidgenössische Technische Hochschule Zürich, Zürich, Switzerland. ✉ email: [email protected]; [email protected] NATURE COMMUNICATIONS | (2021) 12:3053 | https://doi.org/10.1038/s41467-021-23362-y | www.nature.com/naturecommunications 1 ARTICLE NATURE COMMUNICATIONS | https://doi.org/10.1038/s41467-021-23362-y e can program biological systems with DNA that is In previous work, we synthesized and assembled the rewritten Wbased on native nucleotide sequences. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Supplementary Table 3 Complete List of RNA-Sequencing Analysis of Gene Expression Changed by ≥ Tenfold Between Xenograft and Cells Cultured in 10%O2
Supplementary Table 3 Complete list of RNA-Sequencing analysis of gene expression changed by ≥ tenfold between xenograft and cells cultured in 10%O2 Expr Log2 Ratio Symbol Entrez Gene Name (culture/xenograft) -7.182 PGM5 phosphoglucomutase 5 -6.883 GPBAR1 G protein-coupled bile acid receptor 1 -6.683 CPVL carboxypeptidase, vitellogenic like -6.398 MTMR9LP myotubularin related protein 9-like, pseudogene -6.131 SCN7A sodium voltage-gated channel alpha subunit 7 -6.115 POPDC2 popeye domain containing 2 -6.014 LGI1 leucine rich glioma inactivated 1 -5.86 SCN1A sodium voltage-gated channel alpha subunit 1 -5.713 C6 complement C6 -5.365 ANGPTL1 angiopoietin like 1 -5.327 TNN tenascin N -5.228 DHRS2 dehydrogenase/reductase 2 leucine rich repeat and fibronectin type III domain -5.115 LRFN2 containing 2 -5.076 FOXO6 forkhead box O6 -5.035 ETNPPL ethanolamine-phosphate phospho-lyase -4.993 MYO15A myosin XVA -4.972 IGF1 insulin like growth factor 1 -4.956 DLG2 discs large MAGUK scaffold protein 2 -4.86 SCML4 sex comb on midleg like 4 (Drosophila) Src homology 2 domain containing transforming -4.816 SHD protein D -4.764 PLP1 proteolipid protein 1 -4.764 TSPAN32 tetraspanin 32 -4.713 N4BP3 NEDD4 binding protein 3 -4.705 MYOC myocilin -4.646 CLEC3B C-type lectin domain family 3 member B -4.646 C7 complement C7 -4.62 TGM2 transglutaminase 2 -4.562 COL9A1 collagen type IX alpha 1 chain -4.55 SOSTDC1 sclerostin domain containing 1 -4.55 OGN osteoglycin -4.505 DAPL1 death associated protein like 1 -4.491 C10orf105 chromosome 10 open reading frame 105 -4.491 -

Identification of a Mutation in Synapsin I, a Synaptic Vesicle Protein, in a Family with Epilepsy J Med Genet: First Published As on 1 March 2004
183 SHORT REPORT Identification of a mutation in synapsin I, a synaptic vesicle protein, in a family with epilepsy J Med Genet: first published as on 1 March 2004. Downloaded from C C Garcia, H J Blair, M Seager, A Coulthard, S Tennant, M Buddles, A Curtis, J A Goodship ............................................................................................................................... J Med Genet 2004;41:183–187. doi: 10.1136/jmg.2003.013680 hippocampal bodies are moderately atrophic but without A four generation family is described in which some men of signal intensities. normal intelligence have epilepsy and others have various III-2 is a 64 year old man who has had a cerebrovascular combinations of epilepsy, learning difficulties, macrocephaly, accident but was previously of normal intelligence. He had and aggressive behaviour. As the phenotype in this family is tonic-clonic seizures until aged 7. His head circumference is distinct from other X linked recessive disorders linkage studies normal. were carried out. Linkage analysis was done using X III-3 is a 53 year old man with a full scale IQ that has been chromosome microsatellite polymorphisms to define the formally assessed as 72 (with the recognised error in such interval containing the causative gene. Genes from within assessments this places him within the borderline to mild the region were considered possible candidates and one of learning difficulties range). His head circumference is these, SYN1, was screened for mutations by direct DNA normal. As a teenager, he showed extreme physical aggres- sequencing of amplified products. Microsatellite analysis sion (he was held in a high security psychiatric hospital showed that the region between MAOB (Xp11.3) and between the ages of 11 and 18). -

Horizontal Gene Flow Into Geobacillus Is Constrained by the Chromosomal Organization of Growth and Sporulation
bioRxiv preprint doi: https://doi.org/10.1101/381442; this version posted August 2, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Horizontal gene flow into Geobacillus is constrained by the chromosomal organization of growth and sporulation Alexander Esin1,2, Tom Ellis3,4, Tobias Warnecke1,2* 1Molecular Systems Group, Medical Research Council London Institute of Medical Sciences, London, United Kingdom 2Institute of Clinical Sciences, Faculty of Medicine, Imperial College London, London, United Kingdom 3Imperial College Centre for Synthetic Biology, Imperial College London, London, United Kingdom 4Department of Bioengineering, Imperial College London, London, United Kingdom *corresponding author ([email protected]) 1 bioRxiv preprint doi: https://doi.org/10.1101/381442; this version posted August 2, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Abstract Horizontal gene transfer (HGT) in bacteria occurs in the context of adaptive genome architecture. As a consequence, some chromosomal neighbourhoods are likely more permissive to HGT than others. Here, we investigate the chromosomal topology of horizontal gene flow into a clade of Bacillaceae that includes Geobacillus spp. Reconstructing HGT patterns using a phylogenetic approach coupled to model-based reconciliation, we discover three large contiguous chromosomal zones of HGT enrichment. -

Section 4. Guidance Document on Horizontal Gene Transfer Between Bacteria
306 - PART 2. DOCUMENTS ON MICRO-ORGANISMS Section 4. Guidance document on horizontal gene transfer between bacteria 1. Introduction Horizontal gene transfer (HGT) 1 refers to the stable transfer of genetic material from one organism to another without reproduction. The significance of horizontal gene transfer was first recognised when evidence was found for ‘infectious heredity’ of multiple antibiotic resistance to pathogens (Watanabe, 1963). The assumed importance of HGT has changed several times (Doolittle et al., 2003) but there is general agreement now that HGT is a major, if not the dominant, force in bacterial evolution. Massive gene exchanges in completely sequenced genomes were discovered by deviant composition, anomalous phylogenetic distribution, great similarity of genes from distantly related species, and incongruent phylogenetic trees (Ochman et al., 2000; Koonin et al., 2001; Jain et al., 2002; Doolittle et al., 2003; Kurland et al., 2003; Philippe and Douady, 2003). There is also much evidence now for HGT by mobile genetic elements (MGEs) being an ongoing process that plays a primary role in the ecological adaptation of prokaryotes. Well documented is the example of the dissemination of antibiotic resistance genes by HGT that allowed bacterial populations to rapidly adapt to a strong selective pressure by agronomically and medically used antibiotics (Tschäpe, 1994; Witte, 1998; Mazel and Davies, 1999). MGEs shape bacterial genomes, promote intra-species variability and distribute genes between distantly related bacterial genera. Horizontal gene transfer (HGT) between bacteria is driven by three major processes: transformation (the uptake of free DNA), transduction (gene transfer mediated by bacteriophages) and conjugation (gene transfer by means of plasmids or conjugative and integrated elements). -
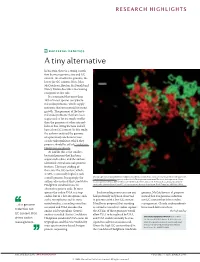
Bacterial Genetics a Tiny Alternative
RESEARCH HIGHLIGHTS BACTERIAL GENETICS A tiny alternative In bacteria, there is a strong correla- tion between genome size and GC content: the smaller the genome, the lower the GC content. Now, John McCutcheon, Bradon McDonald and Nancy Moran describe a fascinating exception to this rule. It is estimated that more than 10% of insect species carry bacte- rial endosymbionts, which supply nutrients that are essential for insect growth. The genomes of the bacte- rial endosymbionts that have been sequenced so far are much smaller than the genomes of other intracel- lular or free-living bacteria and all have a low GC content. In this study, the authors analysed the genome of a previously uncharacterized cicada endosymbiont, which they propose should be called Candidatus Hodgkinia cicadicola. At 144 kb, this is the smallest bacterial genome that has been sequenced to date, and the authors identified several unusual genomic features. The most striking of these was the GC content, which, at 58%, is unusually high for such Micrograph showing Candidatus Hodgkinia cicadicola (red) in close association with another endosymbiont, a small genome. Intriguingly, the Candidatus Sulcia muelleri (green) in the cicada Diceroprocta semicincta. The scale bar represents 10 µm. authors also noticed that Candidatus Image reproduced from McCutcheon, J. P., McDonald, B. R. & Moran, N. P. Origin of an alternative genetic Hodgkinia cicadicola uses an code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 5, e1000565 (2009). alternative genetic code. In most species the codon UGA is a stop Such recoding events are rare and genome, McCutcheon et al. -

Synthetic Biology Projects in Vitro
Downloaded from genome.cshlp.org on September 25, 2021 - Published by Cold Spring Harbor Laboratory Press Review Synthetic biology projects in vitro Anthony C. Forster1,3 and George M. Church2,3 1Department of Pharmacology and Vanderbilt Institute of Chemical Biology, Vanderbilt University Medical Center, Nashville, Tennessee 37232, USA; 2Department of Genetics, Harvard Medical School, Boston, Massachusetts 02115, USA Advances in the in vitro synthesis and evolution of DNA, RNA, and polypeptides are accelerating the construction of biopolymers, pathways, and organisms with novel functions. Known functions are being integrated and debugged with the aim of synthesizing life-like systems. The goals are knowledge, tools, smart materials, and therapies. Synthetic biology projects (SBPs) an action plan to regulate world suppliers of DNA synthesizers, DNA precursors, and oligos (Church 2004). Ethical and safety The basic elements of chemistry and biology are few, but the issues have been, and must continue to be, regulated (Cho et al. synthetic combinations are unlimited and awe inspiring. The 1999). first international conference on synthetic biology charted its Several groups have proposed to create bacteria with chro- goals as understanding and utilizing life’s diverse solutions to mosomes synthesized entirely from synthetic oligos. This might process information, materials, and energy (Silver and Way 2004) be done stepwise (Posfai et al. 2006) or by inactivating the en- (http://syntheticbiology.org). As a bonus, genetic systems are dogenous bacterial chromosome and then somehow transform- biocompatible, renewable, and can be optimized by Darwinian ing and rebooting the bacterium with an entire in vitro- selections. SBPs entail the complex manipulation of replicating synthesized genome. -

Assessment of the Bimodality in the Distribution of Bacterial Genome Sizes
The ISME Journal (2017) 11, 821–824 © 2017 International Society for Microbial Ecology All rights reserved 1751-7362/17 www.nature.com/ismej SHORT COMMUNICATION Assessment of the bimodality in the distribution of bacterial genome sizes Hyun S Gweon, Mark J Bailey and Daniel S Read Centre for Ecology & Hydrology, Wallingford, UK Bacterial genome sizes have previously been shown to exhibit a bimodal distribution. This phenomenon has prompted discussion regarding the evolutionary forces driving genome size in bacteria and its ecological significance. We investigated the level of inherent redundancy in the public database and the effect it has on the shape of the apparent bimodal distribution. Our study reveals that there is a significant bias in the genome sequencing efforts towards a certain group of species, and that correcting the bias using species nomenclature and clustering of the 16S rRNA gene, results in a unimodal rather than the previously published bimodal distribution. The true genome size distribution and its wider ecological implications will soon emerge as we are currently witnessing rapid growth in the number of sequenced genomes from diverse environmental niches across a range of habitats at an unprecedented rate. The ISME Journal (2017) 11, 821–824; doi:10.1038/ismej.2016.142; published online 11 November 2016 Significant progress has been made in understanding Wolf in 2008, where it was reported that bacterial interactions between ecology and genome evolution genome sizes show a bimodal distribution (Koonin in prokaryotes. -

Selective Lowering of Synapsins Induced by Oligomeric Α-Synuclein Exacerbates Memory Deficits
Selective lowering of synapsins induced by oligomeric α-synuclein exacerbates memory deficits Megan E. Larsona,b,c,1, Susan J. Greimela,b,c,1, Fatou Amara,b,c, Michael LaCroixa,b,c, Gabriel Boylea,b,c, Mathew A. Shermana,b,c, Hallie Schleya,b,c, Camille Miela,b,c, Julie A. Schneiderd, Rakez Kayede, Fabio Benfenatif,g, Michael K. Leea,c, David A. Bennettd, and Sylvain E. Lesnéa,b,c,2 aDepartment of Neuroscience, University of Minnesota, Minneapolis, MN 55414; bN. Bud Grossman Center for Memory Research and Care, University of Minnesota, Minneapolis, MN 55414; cInstitute for Translational Neuroscience, University of Minnesota, Minneapolis, MN 55414; dRush Alzheimer’s Disease Center, Rush University Medical Center, Chicago, IL 60612; eDepartment of Neurology, University of Texas Medical Branch, Galveston, TX 77555; fCenter for Synaptic Neuroscience, Istituto Italiano di Tecnologia, 16132 Genoa, Italy; and gDepartment of Experimental Medicine, University of Genova, 16132 Genoa, Italy Edited by Solomon H. Snyder, The Johns Hopkins University School of Medicine, Baltimore, MD, and approved April 24, 2017 (received for review April 4, 2017) Mounting evidence indicates that soluble oligomeric forms of a reduction in the synaptic vesicle recycling pool, and a selective amyloid proteins linked to neurodegenerative disorders, such as lowering of complexins and synapsins (8). Moreover, large mul- amyloid-β (Aβ), tau, or α-synuclein (αSyn) might be the major del- timeric assemblies of recombinant αSyn were recently shown to eterious species for neuronal function in these diseases. Here, we inhibit exocytosis by preferentially binding to synaptobrevin, found an abnormal accumulation of oligomeric αSyn species in AD thereby preventing normal SNARE-mediated vesicle docking (9).