Mechanisms Of, and Barriers To, Horizontal Gene Transfer Between Bacteria
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Transcriptional Landscape of a Rewritten Bacterial Genome Reveals Control Elements and Genome Design Principles ✉ ✉ Mariëlle J
ARTICLE https://doi.org/10.1038/s41467-021-23362-y OPEN The transcriptional landscape of a rewritten bacterial genome reveals control elements and genome design principles ✉ ✉ Mariëlle J. F. M. van Kooten 1 , Clio A. Scheidegger1, Matthias Christen1 & Beat Christen 1 Sequence rewriting enables low-cost genome synthesis and the design of biological systems with orthogonal genetic codes. The error-free, robust rewriting of nucleotide sequences can 1234567890():,; be achieved with a complete annotation of gene regulatory elements. Here, we compare transcription in Caulobacter crescentus to transcription from plasmid-borne segments of the synthesized genome of C. ethensis 2.0. This rewritten derivative contains an extensive amount of supposedly neutral mutations, including 123’562 synonymous codon changes. The tran- scriptional landscape refines 60 promoter annotations, exposes 18 termination elements and links extensive transcription throughout the synthesized genome to the unintentional intro- duction of sigma factor binding motifs. We reveal translational regulation for 20 CDS and uncover an essential translational regulatory element for the expression of ribosomal protein RplS. The annotation of gene regulatory elements allowed us to formulate design principles that improve design schemes for synthesized DNA, en route to a bright future of iteration- free programming of biological systems. 1 Institute of Molecular Systems Biology, Department of Biology, Eidgenössische Technische Hochschule Zürich, Zürich, Switzerland. ✉ email: [email protected]; [email protected] NATURE COMMUNICATIONS | (2021) 12:3053 | https://doi.org/10.1038/s41467-021-23362-y | www.nature.com/naturecommunications 1 ARTICLE NATURE COMMUNICATIONS | https://doi.org/10.1038/s41467-021-23362-y e can program biological systems with DNA that is In previous work, we synthesized and assembled the rewritten Wbased on native nucleotide sequences. -

Laboratory Exercises in Microbiology: Discovering the Unseen World Through Hands-On Investigation
City University of New York (CUNY) CUNY Academic Works Open Educational Resources Queensborough Community College 2016 Laboratory Exercises in Microbiology: Discovering the Unseen World Through Hands-On Investigation Joan Petersen CUNY Queensborough Community College Susan McLaughlin CUNY Queensborough Community College How does access to this work benefit ou?y Let us know! More information about this work at: https://academicworks.cuny.edu/qb_oers/16 Discover additional works at: https://academicworks.cuny.edu This work is made publicly available by the City University of New York (CUNY). Contact: [email protected] Laboratory Exercises in Microbiology: Discovering the Unseen World through Hands-On Investigation By Dr. Susan McLaughlin & Dr. Joan Petersen Queensborough Community College Laboratory Exercises in Microbiology: Discovering the Unseen World through Hands-On Investigation Table of Contents Preface………………………………………………………………………………………i Acknowledgments…………………………………………………………………………..ii Microbiology Lab Safety Instructions…………………………………………………...... iii Lab 1. Introduction to Microscopy and Diversity of Cell Types……………………......... 1 Lab 2. Introduction to Aseptic Techniques and Growth Media………………………...... 19 Lab 3. Preparation of Bacterial Smears and Introduction to Staining…………………...... 37 Lab 4. Acid fast and Endospore Staining……………………………………………......... 49 Lab 5. Metabolic Activities of Bacteria…………………………………………….…....... 59 Lab 6. Dichotomous Keys……………………………………………………………......... 77 Lab 7. The Effect of Physical Factors on Microbial Growth……………………………... 85 Lab 8. Chemical Control of Microbial Growth—Disinfectants and Antibiotics…………. 99 Lab 9. The Microbiology of Milk and Food………………………………………………. 111 Lab 10. The Eukaryotes………………………………………………………………........ 123 Lab 11. Clinical Microbiology I; Anaerobic pathogens; Vectors of Infectious Disease….. 141 Lab 12. Clinical Microbiology II—Immunology and the Biolog System………………… 153 Lab 13. Putting it all Together: Case Studies in Microbiology…………………………… 163 Appendix I. -

Potential Probiotic Bacillus Subtilis Isolated from a Novel Niche
microorganisms Article Potential Probiotic Bacillus subtilis Isolated from a Novel Niche Exhibits Broad Range Antibacterial Activity and Causes Virulence and Metabolic Dysregulation in Enterotoxic E. coli Sudhanshu Sudan 1, Robert Flick 2, Linda Nong 3 and Julang Li 1,* 1 Department of Animal Biosciences, University of Guelph, Guelph, ON N1G 2W1, Canada; [email protected] 2 Biozone, Mass Spectrometry and Metabolomics, Department of Chemical Engineering and Applied Chemistry, University of Toronto, Toronto, ON M5S 3E5, Canada; robert.fl[email protected] 3 Department of Integrative Biology, University of Guelph, Guelph, ON N1G 2W1, Canada; [email protected] * Correspondence: [email protected] Abstract: Microbial life in extreme environments, such as deserts and deep oceans, is thought to have evolved to overcome constraints of nutrient availability, temperature, and suboptimal hygiene environments. Isolation of probiotic bacteria from such niche may provide a competitive edge over traditional probiotics. Here, we tested the survival, safety, and antimicrobial effect of a recently isolated and potential novel strain of Bacillus subtilis (CP9) from desert camel in vitro. Antimicrobial assays were performed via radial diffusion, agar spot, and co-culture assays. Cytotoxic analysis was Citation: Sudan, S.; Flick, R.; Nong, performed using pig intestinal epithelial cells (IPEC-J2). Real time-PCR was performed for studying L.; Li, J. Potential Probiotic Bacillus the effect on ETEC virulence genes and metabolomic analysis was performed using LC-MS. The subtilis Isolated from a Novel Niche results showed that CP9 cells were viable in varied bile salts and in low pH environments. CP9 Exhibits Broad Range Antibacterial showed no apparent cytotoxicity in IPEC-J2 cells. -

Arxiv.Org | Cornell University Library, July, 2019. 1
arXiv.org | Cornell University Library, July, 2019. Extremophiles: a special or general case in the search for extra-terrestrial life? Ian von Hegner Aarhus University Abstract Since time immemorial life has been viewed as fragile, yet over the past few decades it has been found that many extreme environments are inhabited by organisms known as extremophiles. Knowledge of their emergence, adaptability, and limitations seems to provide a guideline for the search of extra-terrestrial life, since some extremophiles presumably can survive in extreme environments such as Mars, Europa, and Enceladus. Due to physico-chemical constraints, the first life necessarily came into existence at the lower limit of it‟s conceivable complexity. Thus, the first life could not have been an extremophile, furthermore, since biological evolution occurs over time, then the dual knowledge regarding what specific extremophiles are capable of, and to the analogue environment on extreme worlds, will not be sufficient as a search criterion. This is because, even though an extremophile can live in an extreme environment here-and-now, its ancestor however could not live in that very same environment in the past, which means that no contemporary extremophiles exist in that environment. Furthermore, a theoretical framework should be able to predict whether extremophiles can be considered a special or general case in the galaxy. Thus, a question is raised: does Earth‟s continuous habitability represent an extreme or average value for planets? Thus, dependent on whether it is difficult or easy for worlds to maintain the habitability, the search for extra- terrestrial life with a focus on extremophiles will either represent a search for dying worlds, or a search for special life on living worlds, focusing too narrowly on extreme values. -

Horizontal Gene Flow Into Geobacillus Is Constrained by the Chromosomal Organization of Growth and Sporulation
bioRxiv preprint doi: https://doi.org/10.1101/381442; this version posted August 2, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Horizontal gene flow into Geobacillus is constrained by the chromosomal organization of growth and sporulation Alexander Esin1,2, Tom Ellis3,4, Tobias Warnecke1,2* 1Molecular Systems Group, Medical Research Council London Institute of Medical Sciences, London, United Kingdom 2Institute of Clinical Sciences, Faculty of Medicine, Imperial College London, London, United Kingdom 3Imperial College Centre for Synthetic Biology, Imperial College London, London, United Kingdom 4Department of Bioengineering, Imperial College London, London, United Kingdom *corresponding author ([email protected]) 1 bioRxiv preprint doi: https://doi.org/10.1101/381442; this version posted August 2, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Abstract Horizontal gene transfer (HGT) in bacteria occurs in the context of adaptive genome architecture. As a consequence, some chromosomal neighbourhoods are likely more permissive to HGT than others. Here, we investigate the chromosomal topology of horizontal gene flow into a clade of Bacillaceae that includes Geobacillus spp. Reconstructing HGT patterns using a phylogenetic approach coupled to model-based reconciliation, we discover three large contiguous chromosomal zones of HGT enrichment. -

Section 4. Guidance Document on Horizontal Gene Transfer Between Bacteria
306 - PART 2. DOCUMENTS ON MICRO-ORGANISMS Section 4. Guidance document on horizontal gene transfer between bacteria 1. Introduction Horizontal gene transfer (HGT) 1 refers to the stable transfer of genetic material from one organism to another without reproduction. The significance of horizontal gene transfer was first recognised when evidence was found for ‘infectious heredity’ of multiple antibiotic resistance to pathogens (Watanabe, 1963). The assumed importance of HGT has changed several times (Doolittle et al., 2003) but there is general agreement now that HGT is a major, if not the dominant, force in bacterial evolution. Massive gene exchanges in completely sequenced genomes were discovered by deviant composition, anomalous phylogenetic distribution, great similarity of genes from distantly related species, and incongruent phylogenetic trees (Ochman et al., 2000; Koonin et al., 2001; Jain et al., 2002; Doolittle et al., 2003; Kurland et al., 2003; Philippe and Douady, 2003). There is also much evidence now for HGT by mobile genetic elements (MGEs) being an ongoing process that plays a primary role in the ecological adaptation of prokaryotes. Well documented is the example of the dissemination of antibiotic resistance genes by HGT that allowed bacterial populations to rapidly adapt to a strong selective pressure by agronomically and medically used antibiotics (Tschäpe, 1994; Witte, 1998; Mazel and Davies, 1999). MGEs shape bacterial genomes, promote intra-species variability and distribute genes between distantly related bacterial genera. Horizontal gene transfer (HGT) between bacteria is driven by three major processes: transformation (the uptake of free DNA), transduction (gene transfer mediated by bacteriophages) and conjugation (gene transfer by means of plasmids or conjugative and integrated elements). -

Arsenite Efflux Limits Microbial Arsenic Volatilization
Arsenite eux limits microbial arsenic volatilization Changdong Ke Huaqiao University Qingyue Kuang Huaqiao University Chungui Zhao ( [email protected] ) Jiafu Luo Huaqiao University Wenzhang Wei Huaqiao University Hend A. Alwathnani King Suad University Quaiser Saquib King Saud University Yanshuang Yu King Suad University Christopher Rensing Fujian Agriculture and Forestry University Suping Yang Huaqiao University Research article Keywords: Arsenic, As eux transporter, As methylation, Rhodopseudomonas palustris Posted Date: July 4th, 2019 DOI: https://doi.org/10.21203/rs.2.10508/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/17 Abstract Background: Arsenic (As) methylation is regarded as a potential way to volatize and thereby remove As from the environment. However, most microorganisms conducting As methylation display low As volatilization eciency as As methylation is limited by As eux transporters as both processes compete for arsenite [As(III)]. In the study, we deleted arsB and acr3 from Rhodopseudomonas palustris CGA009, a good model organism for studying As detoxication, and further investigated the effect of As(III) eux transporters on As methylation. Results: Two mutants were obtained by gene deletion. Compared to the growth inhibition rate (IC50) [1.57±0.11 mmol/L As(III) and 2.67±0.04 mmol/L arsenate [As(V)] of wildtype R. palustris CGA009, the As(III) and As(V) resistance of the mutants decreased, and IC50 value of the R. palustris CGA009 ∆arsB mutant was 1.47±0.02 mmol/L As(III) and 2.12±0.03 mmol/L As(V), respectively, and that of the R. -
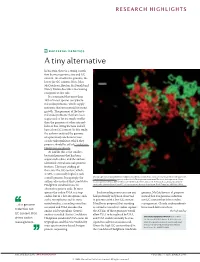
Bacterial Genetics a Tiny Alternative
RESEARCH HIGHLIGHTS BACTERIAL GENETICS A tiny alternative In bacteria, there is a strong correla- tion between genome size and GC content: the smaller the genome, the lower the GC content. Now, John McCutcheon, Bradon McDonald and Nancy Moran describe a fascinating exception to this rule. It is estimated that more than 10% of insect species carry bacte- rial endosymbionts, which supply nutrients that are essential for insect growth. The genomes of the bacte- rial endosymbionts that have been sequenced so far are much smaller than the genomes of other intracel- lular or free-living bacteria and all have a low GC content. In this study, the authors analysed the genome of a previously uncharacterized cicada endosymbiont, which they propose should be called Candidatus Hodgkinia cicadicola. At 144 kb, this is the smallest bacterial genome that has been sequenced to date, and the authors identified several unusual genomic features. The most striking of these was the GC content, which, at 58%, is unusually high for such Micrograph showing Candidatus Hodgkinia cicadicola (red) in close association with another endosymbiont, a small genome. Intriguingly, the Candidatus Sulcia muelleri (green) in the cicada Diceroprocta semicincta. The scale bar represents 10 µm. authors also noticed that Candidatus Image reproduced from McCutcheon, J. P., McDonald, B. R. & Moran, N. P. Origin of an alternative genetic Hodgkinia cicadicola uses an code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 5, e1000565 (2009). alternative genetic code. In most species the codon UGA is a stop Such recoding events are rare and genome, McCutcheon et al. -

Synthetic Biology Projects in Vitro
Downloaded from genome.cshlp.org on September 25, 2021 - Published by Cold Spring Harbor Laboratory Press Review Synthetic biology projects in vitro Anthony C. Forster1,3 and George M. Church2,3 1Department of Pharmacology and Vanderbilt Institute of Chemical Biology, Vanderbilt University Medical Center, Nashville, Tennessee 37232, USA; 2Department of Genetics, Harvard Medical School, Boston, Massachusetts 02115, USA Advances in the in vitro synthesis and evolution of DNA, RNA, and polypeptides are accelerating the construction of biopolymers, pathways, and organisms with novel functions. Known functions are being integrated and debugged with the aim of synthesizing life-like systems. The goals are knowledge, tools, smart materials, and therapies. Synthetic biology projects (SBPs) an action plan to regulate world suppliers of DNA synthesizers, DNA precursors, and oligos (Church 2004). Ethical and safety The basic elements of chemistry and biology are few, but the issues have been, and must continue to be, regulated (Cho et al. synthetic combinations are unlimited and awe inspiring. The 1999). first international conference on synthetic biology charted its Several groups have proposed to create bacteria with chro- goals as understanding and utilizing life’s diverse solutions to mosomes synthesized entirely from synthetic oligos. This might process information, materials, and energy (Silver and Way 2004) be done stepwise (Posfai et al. 2006) or by inactivating the en- (http://syntheticbiology.org). As a bonus, genetic systems are dogenous bacterial chromosome and then somehow transform- biocompatible, renewable, and can be optimized by Darwinian ing and rebooting the bacterium with an entire in vitro- selections. SBPs entail the complex manipulation of replicating synthesized genome. -

Assessment of the Bimodality in the Distribution of Bacterial Genome Sizes
The ISME Journal (2017) 11, 821–824 © 2017 International Society for Microbial Ecology All rights reserved 1751-7362/17 www.nature.com/ismej SHORT COMMUNICATION Assessment of the bimodality in the distribution of bacterial genome sizes Hyun S Gweon, Mark J Bailey and Daniel S Read Centre for Ecology & Hydrology, Wallingford, UK Bacterial genome sizes have previously been shown to exhibit a bimodal distribution. This phenomenon has prompted discussion regarding the evolutionary forces driving genome size in bacteria and its ecological significance. We investigated the level of inherent redundancy in the public database and the effect it has on the shape of the apparent bimodal distribution. Our study reveals that there is a significant bias in the genome sequencing efforts towards a certain group of species, and that correcting the bias using species nomenclature and clustering of the 16S rRNA gene, results in a unimodal rather than the previously published bimodal distribution. The true genome size distribution and its wider ecological implications will soon emerge as we are currently witnessing rapid growth in the number of sequenced genomes from diverse environmental niches across a range of habitats at an unprecedented rate. The ISME Journal (2017) 11, 821–824; doi:10.1038/ismej.2016.142; published online 11 November 2016 Significant progress has been made in understanding Wolf in 2008, where it was reported that bacterial interactions between ecology and genome evolution genome sizes show a bimodal distribution (Koonin in prokaryotes. -

Selection and Robustness in Bacterial Genome Evolution Seila Omer University of Connecticut, [email protected]
University of Connecticut OpenCommons@UConn Doctoral Dissertations University of Connecticut Graduate School 12-16-2016 Selection and Robustness in Bacterial Genome Evolution Seila Omer University Of Connecticut, [email protected] Follow this and additional works at: https://opencommons.uconn.edu/dissertations Recommended Citation Omer, Seila, "Selection and Robustness in Bacterial Genome Evolution" (2016). Doctoral Dissertations. 1317. https://opencommons.uconn.edu/dissertations/1317 Selection and Robustness in Bacterial Genome Evolution Seila Omer, Ph.D. University of Connecticut, 2016 The research presented in this thesis attempts to address research questions related to the role of natural selection in the evolution of bacterial genes not expressed for function and in building mutational tolerance to translational errors. Studies on evolution of protein coding DNA sequences have provided the evidence for a current paradigm in evolutionary biology: only functional genes are undergoing selection against the deleterious effects of allele variants (purifying selection). I provide evidence that similar footprints of selection can be detected in genes that are not normally expressed for function during the bacterial life cycle. Using simulations for DNA sequence evolution, I demonstrate statistically significant deviations from neutral evolution for the studied genes. I suggest that purifying selection affects both functional and non-functional genes. I propose this might be caused by the dominant toxic effects of low level translation of mutated products in bacteria, due to misfolding and misinteraction. Natural selection also acts to remove the effects of translational errors. Stop codon readthrough events are more likely to have major structural and functional effects than simple nucleotide changes. Recent research has shown that strength of selection experienced by protein-coding genes is positively correlated with the level of gene expression. -

Bacterial Transformation Buffers Environmental Fluctuations Through the Reversible Integration of Mobile Genetic Elements
bioRxiv preprint doi: https://doi.org/10.1101/557462; this version posted February 24, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Bacterial transformation buffers environmental fluctuations through the reversible integration of mobile genetic elements Gabriel Carvalho1, David Fouchet1, Gonché Danesh1, Anne-Sophie Godeux2, Maria-Halima Laaberki2,3, Dominique Pontier1, Xavier Charpentier2, 4,*, Samuel Venner1, 4, * 1Laboratoire de Biométrie et Biologie Evolutive, CNRS UMR5558, Université Claude Bernard Lyon 1, Villeurbanne, France 2CIRI, Centre International de Recherche en Infectiologie, Inserm, U1111, Université Claude Bernard Lyon 1, CNRS, UMR5308, École Normale Supérieure de Lyon, Univ Lyon, 69100, Villeurbanne, France 3Université de Lyon, VetAgro Sup, 69280 Marcy l'Etoile, France. 4 These authors contributed equally to this study * Corresponding authors: [email protected], [email protected] Abstract Horizontal gene transfer (HGT) is known to promote the spread of genes in bacterial communities, which is of primary importance to human health when these genes provide resistance to antibiotics. Among the main HGT mechanisms, natural transformation stands out as being widespread and encoded by the bacterial core genome. From an evolutionary perspective, transformation is often viewed as a mean to generate genetic diversity and mixing within bacterial populations. However, another recent paradigm proposes that its main evolutionary function would be to cure bacterial genomes from their parasitic mobile genetic elements (MGEs). Here, we propose to combine these two seemingly opposing points of view because MGEs, although costly for bacterial cells, can carry functions that are point-in-time beneficial to bacteria under stressful conditions (e.g.