Insights from 20 Years of Bacterial Genome Sequencing
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Transcriptional Landscape of a Rewritten Bacterial Genome Reveals Control Elements and Genome Design Principles ✉ ✉ Mariëlle J
ARTICLE https://doi.org/10.1038/s41467-021-23362-y OPEN The transcriptional landscape of a rewritten bacterial genome reveals control elements and genome design principles ✉ ✉ Mariëlle J. F. M. van Kooten 1 , Clio A. Scheidegger1, Matthias Christen1 & Beat Christen 1 Sequence rewriting enables low-cost genome synthesis and the design of biological systems with orthogonal genetic codes. The error-free, robust rewriting of nucleotide sequences can 1234567890():,; be achieved with a complete annotation of gene regulatory elements. Here, we compare transcription in Caulobacter crescentus to transcription from plasmid-borne segments of the synthesized genome of C. ethensis 2.0. This rewritten derivative contains an extensive amount of supposedly neutral mutations, including 123’562 synonymous codon changes. The tran- scriptional landscape refines 60 promoter annotations, exposes 18 termination elements and links extensive transcription throughout the synthesized genome to the unintentional intro- duction of sigma factor binding motifs. We reveal translational regulation for 20 CDS and uncover an essential translational regulatory element for the expression of ribosomal protein RplS. The annotation of gene regulatory elements allowed us to formulate design principles that improve design schemes for synthesized DNA, en route to a bright future of iteration- free programming of biological systems. 1 Institute of Molecular Systems Biology, Department of Biology, Eidgenössische Technische Hochschule Zürich, Zürich, Switzerland. ✉ email: [email protected]; [email protected] NATURE COMMUNICATIONS | (2021) 12:3053 | https://doi.org/10.1038/s41467-021-23362-y | www.nature.com/naturecommunications 1 ARTICLE NATURE COMMUNICATIONS | https://doi.org/10.1038/s41467-021-23362-y e can program biological systems with DNA that is In previous work, we synthesized and assembled the rewritten Wbased on native nucleotide sequences. -

Robust Taxonomic Classification of Uncharted Microbial Sequences and Bins with CAT and BAT
bioRxiv preprint doi: https://doi.org/10.1101/530188; this version posted January 24, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. Robust taxonomic classification of uncharted microbial sequences and bins with CAT and BAT F.A. Bastiaan von Meijenfeldt1,†, Ksenia Arkhipova1,†, Diego D. Cambuy1, Felipe H. Coutinho2,3, Bas E. Dutilh1,2,* 1 Theoretical Biology and Bioinformatics, Science for Life, Utrecht University, The Netherlands. 2 Centre for Molecular and Biomolecular Informatics, Radboud University Medical Centre, Nijmegen, The Netherlands. 3 Instituto de Biologia, Universidade Federal do Rio de Janeiro, Rio de Janeiro, RJ, Brazil. * To whom correspondence should be addressed. Tel: +31 30 253 4212; Email: [email protected]. † These authors contributed equally to this work. Present Address: [Felipe H. Couthinho], Evolutionary Genomics Group, Departamento de Produccíon y Microbiología, Universidad Miguel Hernández, Campus San Juan, San Juan, Alicante 03550, Spain. ABSTRACT Current-day metagenomics increasingly requires taxonomic classification of long DNA sequences and metagenome-assembled genomes (MAGs) of unknown microorganisms. We show that the standard best-hit approach often leads to classifications that are too specific. We present tools to classify high- quality metagenomic contigs (Contig Annotation Tool, CAT) and MAGs (Bin Annotation Tool, BAT) and thoroughly benchmark them with simulated metagenomic sequences that are classified against a reference database where related sequences are increasingly removed, thereby simulating increasingly unknown queries. We find that the query sequences are correctly classified at low taxonomic ranks if closely related organisms are present in the reference database, while classifications are made higher in the taxonomy when closely related organisms are absent, thus avoiding spurious classification specificity. -

First Genomic Insights Into Members of a Candidate Bacterial Phylum Responsible for Wastewater Bulking
First genomic insights into members of a candidate bacterial phylum responsible for wastewater bulking Yuji Sekiguchi1, Akiko Ohashi1, Donovan H. Parks2, Toshihiro Yamauchi3, Gene W. Tyson2,4 and Philip Hugenholtz2,5 1 Biomedical Research Institute, National Institute of Advanced Industrial Science and Technology (AIST), Tsukuba, Ibaraki, Japan 2 Australian Centre for Ecogenomics, School of Chemistry and Molecular Biosciences, The University of Queensland, St. Lucia, Queensland, Australia 3 Administrative Management Department, Kubota Kasui Corporation, Minato-ku, Tokyo, Japan 4 Advanced Water Management Centre, The University of Queensland, St. Lucia, Queensland, Australia 5 Institute for Molecular Bioscience, The University of Queensland, St. Lucia, Queensland, Australia ABSTRACT Filamentous cells belonging to the candidate bacterial phylum KSB3 were previously identified as the causative agent of fatal filament overgrowth (bulking) in a high-rate industrial anaerobic wastewater treatment bioreactor. Here, we obtained near complete genomes from two KSB3 populations in the bioreactor, including the dominant bulking filament, using diVerential coverage binning of metagenomic data. Fluorescence in situ hybridization with 16S rRNA-targeted probes specific for the two populations confirmed that both are filamentous organisms. Genome-based metabolic reconstruction and microscopic observation of the KSB3 filaments in the presence of sugar gradients indicate that both filament types are Gram-negative, strictly anaerobic fermenters capable of -

Metal Transformation by a Novel Pelosinus Isolate from a Subsurface Environment
INL/JOU-08-14091-Revision-0 Metal Transformation by a Novel Pelosinus Isolate From a Subsurface Environment Allison E. Ray, Peter P. Sheridan, Andrew L. Neal, Yoshiko Fujita, David E. Cummings, Timothy S. Magnuson August 2018 The INL is a U.S. Department of Energy National Laboratory operated by Battelle Energy Alliance INL/JOU-08-14091-Revision-0 Metal Transformation by a Novel Pelosinus Isolate From a Subsurface Environment Allison E. Ray, Peter P. Sheridan, Andrew L. Neal, Yoshiko Fujita, David E. Cummings, Timothy S. Magnuson August 2018 Idaho National Laboratory Idaho Falls, Idaho 83415 http://www.inl.gov Prepared for the U.S. Department of Energy Office of Nuclear Energy Under DOE Idaho Operations Office Contract DE-AC07-05ID14517 fmicb-09-01689 July 25, 2018 Time: 20:42 # 1 ORIGINAL RESEARCH published: xx July 2018 doi: 10.3389/fmicb.2018.01689 1 58 2 59 3 60 4 61 5 62 6 63 7 64 8 65 9 Metal Transformation by a Novel 66 10 67 11 Pelosinus Isolate From a Subsurface 68 12 69 13 Environment 70 14 71 15 Allison E. Ray1,2, Stephanie A. Connon1,3, Andrew L. Neal4†, Yoshiko Fujita2, 72 5 2† 1 16 David E. Cummings , Jani C. Ingram and Timothy S. Magnuson * 73 17 74 1 Department of Biological Sciences, Idaho State University, Pocatello, ID, United States, 2 Bioenergy Technologies, Idaho 18 3 75 Edited by: National Laboratory, Idaho Falls, ID, United States, California Institute of Technology, Pasadena, CA, United States, 19 4 5 76 Pankaj Kumar Arora, Savannah River Ecology Laboratory, University of Georgia, Aiken, SC, United States, Department of Biology, Point Loma 20 77 Babasaheb Bhimrao Ambedkar Nazarene University, San Diego, CA, United States 21 University, India 78 22 79 Reviewed by: The capability of microorganisms to alter metal speciation offers potential for 23 80 Ramprasad E.V.V., the development of new strategies for immobilization of toxic metals in the 24 University of Hyderabad, India 81 25 Bärbel Ulrike Fösel, environment. -

Horizontal Gene Flow Into Geobacillus Is Constrained by the Chromosomal Organization of Growth and Sporulation
bioRxiv preprint doi: https://doi.org/10.1101/381442; this version posted August 2, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Horizontal gene flow into Geobacillus is constrained by the chromosomal organization of growth and sporulation Alexander Esin1,2, Tom Ellis3,4, Tobias Warnecke1,2* 1Molecular Systems Group, Medical Research Council London Institute of Medical Sciences, London, United Kingdom 2Institute of Clinical Sciences, Faculty of Medicine, Imperial College London, London, United Kingdom 3Imperial College Centre for Synthetic Biology, Imperial College London, London, United Kingdom 4Department of Bioengineering, Imperial College London, London, United Kingdom *corresponding author ([email protected]) 1 bioRxiv preprint doi: https://doi.org/10.1101/381442; this version posted August 2, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Abstract Horizontal gene transfer (HGT) in bacteria occurs in the context of adaptive genome architecture. As a consequence, some chromosomal neighbourhoods are likely more permissive to HGT than others. Here, we investigate the chromosomal topology of horizontal gene flow into a clade of Bacillaceae that includes Geobacillus spp. Reconstructing HGT patterns using a phylogenetic approach coupled to model-based reconciliation, we discover three large contiguous chromosomal zones of HGT enrichment. -

Section 4. Guidance Document on Horizontal Gene Transfer Between Bacteria
306 - PART 2. DOCUMENTS ON MICRO-ORGANISMS Section 4. Guidance document on horizontal gene transfer between bacteria 1. Introduction Horizontal gene transfer (HGT) 1 refers to the stable transfer of genetic material from one organism to another without reproduction. The significance of horizontal gene transfer was first recognised when evidence was found for ‘infectious heredity’ of multiple antibiotic resistance to pathogens (Watanabe, 1963). The assumed importance of HGT has changed several times (Doolittle et al., 2003) but there is general agreement now that HGT is a major, if not the dominant, force in bacterial evolution. Massive gene exchanges in completely sequenced genomes were discovered by deviant composition, anomalous phylogenetic distribution, great similarity of genes from distantly related species, and incongruent phylogenetic trees (Ochman et al., 2000; Koonin et al., 2001; Jain et al., 2002; Doolittle et al., 2003; Kurland et al., 2003; Philippe and Douady, 2003). There is also much evidence now for HGT by mobile genetic elements (MGEs) being an ongoing process that plays a primary role in the ecological adaptation of prokaryotes. Well documented is the example of the dissemination of antibiotic resistance genes by HGT that allowed bacterial populations to rapidly adapt to a strong selective pressure by agronomically and medically used antibiotics (Tschäpe, 1994; Witte, 1998; Mazel and Davies, 1999). MGEs shape bacterial genomes, promote intra-species variability and distribute genes between distantly related bacterial genera. Horizontal gene transfer (HGT) between bacteria is driven by three major processes: transformation (the uptake of free DNA), transduction (gene transfer mediated by bacteriophages) and conjugation (gene transfer by means of plasmids or conjugative and integrated elements). -
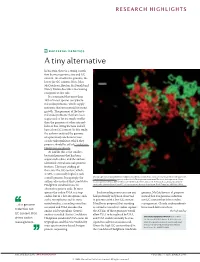
Bacterial Genetics a Tiny Alternative
RESEARCH HIGHLIGHTS BACTERIAL GENETICS A tiny alternative In bacteria, there is a strong correla- tion between genome size and GC content: the smaller the genome, the lower the GC content. Now, John McCutcheon, Bradon McDonald and Nancy Moran describe a fascinating exception to this rule. It is estimated that more than 10% of insect species carry bacte- rial endosymbionts, which supply nutrients that are essential for insect growth. The genomes of the bacte- rial endosymbionts that have been sequenced so far are much smaller than the genomes of other intracel- lular or free-living bacteria and all have a low GC content. In this study, the authors analysed the genome of a previously uncharacterized cicada endosymbiont, which they propose should be called Candidatus Hodgkinia cicadicola. At 144 kb, this is the smallest bacterial genome that has been sequenced to date, and the authors identified several unusual genomic features. The most striking of these was the GC content, which, at 58%, is unusually high for such Micrograph showing Candidatus Hodgkinia cicadicola (red) in close association with another endosymbiont, a small genome. Intriguingly, the Candidatus Sulcia muelleri (green) in the cicada Diceroprocta semicincta. The scale bar represents 10 µm. authors also noticed that Candidatus Image reproduced from McCutcheon, J. P., McDonald, B. R. & Moran, N. P. Origin of an alternative genetic Hodgkinia cicadicola uses an code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 5, e1000565 (2009). alternative genetic code. In most species the codon UGA is a stop Such recoding events are rare and genome, McCutcheon et al. -

Synthetic Biology Projects in Vitro
Downloaded from genome.cshlp.org on September 25, 2021 - Published by Cold Spring Harbor Laboratory Press Review Synthetic biology projects in vitro Anthony C. Forster1,3 and George M. Church2,3 1Department of Pharmacology and Vanderbilt Institute of Chemical Biology, Vanderbilt University Medical Center, Nashville, Tennessee 37232, USA; 2Department of Genetics, Harvard Medical School, Boston, Massachusetts 02115, USA Advances in the in vitro synthesis and evolution of DNA, RNA, and polypeptides are accelerating the construction of biopolymers, pathways, and organisms with novel functions. Known functions are being integrated and debugged with the aim of synthesizing life-like systems. The goals are knowledge, tools, smart materials, and therapies. Synthetic biology projects (SBPs) an action plan to regulate world suppliers of DNA synthesizers, DNA precursors, and oligos (Church 2004). Ethical and safety The basic elements of chemistry and biology are few, but the issues have been, and must continue to be, regulated (Cho et al. synthetic combinations are unlimited and awe inspiring. The 1999). first international conference on synthetic biology charted its Several groups have proposed to create bacteria with chro- goals as understanding and utilizing life’s diverse solutions to mosomes synthesized entirely from synthetic oligos. This might process information, materials, and energy (Silver and Way 2004) be done stepwise (Posfai et al. 2006) or by inactivating the en- (http://syntheticbiology.org). As a bonus, genetic systems are dogenous bacterial chromosome and then somehow transform- biocompatible, renewable, and can be optimized by Darwinian ing and rebooting the bacterium with an entire in vitro- selections. SBPs entail the complex manipulation of replicating synthesized genome. -

Sensitive and Specific Detection of Pseudomonas Avellanae Using
J. Phytopathology 149, 527±532 (2001) Ó 2001 Blackwell Wissenschafts-Verlag, Berlin ISSN 0931-1785 Istituto Sperimentale per la Frutticoltura, Ciampino Aeroporto, Roma, Italy Sensitive and Speci®c Detection of Pseudomonas avellanae using Primers based on 16S rRNA Gene Sequences M. SCORTICHINI* and U. MARCHESI Authors' address: Istituto Sperimentale per la Frutticoltura, Via di Fioranello, 52, I-00040 Ciampino Aeroporto, Rome, Italy (correspondence to M. Scortichini. E-mail: [email protected]) With 4 ®gures Received January 1, 2001; accepted May 1, 2001 Keywords: Pseudomonas avellanae, 16S rRNA gene, detection, hazelnut decline, primers Abstract up to some years. The detection of P. avellanae is A rapid polymerase chain reaction (PCR)-based proce- currently based either on traditional techniques that dure was developed for the detection of Pseudomonas include pathogenicity tests which require at least avellanae, the causal agent of hazelnut (Corylus avellana) 6±7 months for the completion or on repetitive-PCR decline in northern Greece and central Italy. The partial using the ERIC primers and requiring the isolation and sequence of the 16S rRNA gene of P. avellanae strain the production of pure cultures (Scortichini et al., PD 2390, isolated in central Italy, was compared with 2000a). The latter procedure can be completed in the sequence coding for the same gene of P. syringae pv. 4±6 days but latent infection cannot be detected. The syringae type-strain LMG 1247t1. Primers PAV 1 and possibility of utilizing a diagnostic technique enabling a PAV 22 were chosen, and after the PCR, an ampli®ca- reliable and rapid screening of the propagative material, tion product of 762 base pairs was speci®cally produced can also support the establishing of new hazelnut only by 40 strains of P. -

EXPERIMENTAL STUDIES on FERMENTATIVE FIRMICUTES from ANOXIC ENVIRONMENTS: ISOLATION, EVOLUTION, and THEIR GEOCHEMICAL IMPACTS By
EXPERIMENTAL STUDIES ON FERMENTATIVE FIRMICUTES FROM ANOXIC ENVIRONMENTS: ISOLATION, EVOLUTION, AND THEIR GEOCHEMICAL IMPACTS By JESSICA KEE EUN CHOI A dissertation submitted to the School of Graduate Studies Rutgers, The State University of New Jersey In partial fulfillment of the requirements For the degree of Doctor of Philosophy Graduate Program in Microbial Biology Written under the direction of Nathan Yee And approved by _______________________________________________________ _______________________________________________________ _______________________________________________________ _______________________________________________________ New Brunswick, New Jersey October 2017 ABSTRACT OF THE DISSERTATION Experimental studies on fermentative Firmicutes from anoxic environments: isolation, evolution and their geochemical impacts by JESSICA KEE EUN CHOI Dissertation director: Nathan Yee Fermentative microorganisms from the bacterial phylum Firmicutes are quite ubiquitous in subsurface environments and play an important biogeochemical role. For instance, fermenters have the ability to take complex molecules and break them into simpler compounds that serve as growth substrates for other organisms. The research presented here focuses on two groups of fermentative Firmicutes, one from the genus Clostridium and the other from the class Negativicutes. Clostridium species are well-known fermenters. Laboratory studies done so far have also displayed the capability to reduce Fe(III), yet the mechanism of this activity has not been investigated -

Supplementary Figure Legends for Rands Et Al. 2019
Supplementary Figure legends for Rands et al. 2019 Figure S1: Display of all 485 prophage genome maps predicted from Gram-Negative Firmicutes. Each horizontal line corresponds to an individual prophage shown to scale and color-coded for annotated phage genes according to the key displayed in the right- side Box. The left vertical Bar indicates the Bacterial host in a colour code. Figure S2: Projection of virome sequences from 183 human stool samples on A. Acidaminococcus intestini RYC-MR95, and B. Veillonella parvula UTDB1-3. The first panel shows the read coverage (Y-axis) across the complete Bacterial genome sequence (X-axis; with bp coordinates). Predicted prophage regions are marked with red triangles and magnified in the suBsequent panels. Virome reads projected outside of prophage prediction are listed in Table S4. Figure S3: The same display of virome sequences projected onto Bacterial genomes as in Figure S2, But for two different Negativicute species: A. Dialister Marseille, and B. Negativicoccus massiliensis. For non-phage peak annotations, see Table S4. Figure S4: Gene flanking analysis for the lysis module from all prophages predicted in all the different Bacterial clades (Table S2), a total of 3,462 prophages. The lysis module is generally located next to the tail module in Firmicute prophages, But adjacent to the packaging (terminase) module in Escherichia phages. 1 Figure S5: Candidate Mu-like prophage in the Negativicute Propionispora vibrioides. Phage-related genes (arrows indicate transcription direction) are coloured and show characteristics of Mu-like genome organization. Figure S6: The genome maps of Negativicute prophages harbouring candidate antiBiotic resistance genes MBL (top three Veillonella prophages) and tet(32) (bottom Selenomonas prophage remnant); excludes the ACI-1 prophage harbouring example characterised previously (Rands et al., 2018). -

International Journal of Systematic and Evolutionary Microbiology (2016), 66, 5575–5599 DOI 10.1099/Ijsem.0.001485
International Journal of Systematic and Evolutionary Microbiology (2016), 66, 5575–5599 DOI 10.1099/ijsem.0.001485 Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Mobolaji Adeolu,† Seema Alnajar,† Sohail Naushad and Radhey S. Gupta Correspondence Department of Biochemistry and Biomedical Sciences, McMaster University, Hamilton, Ontario, Radhey S. Gupta L8N 3Z5, Canada [email protected] Understanding of the phylogeny and interrelationships of the genera within the order ‘Enterobacteriales’ has proven difficult using the 16S rRNA gene and other single-gene or limited multi-gene approaches. In this work, we have completed comprehensive comparative genomic analyses of the members of the order ‘Enterobacteriales’ which includes phylogenetic reconstructions based on 1548 core proteins, 53 ribosomal proteins and four multilocus sequence analysis proteins, as well as examining the overall genome similarity amongst the members of this order. The results of these analyses all support the existence of seven distinct monophyletic groups of genera within the order ‘Enterobacteriales’. In parallel, our analyses of protein sequences from the ‘Enterobacteriales’ genomes have identified numerous molecular characteristics in the forms of conserved signature insertions/deletions, which are specifically shared by the members of the identified clades and independently support their monophyly and distinctness. Many of these groupings, either in part or in whole, have been recognized in previous evolutionary studies, but have not been consistently resolved as monophyletic entities in 16S rRNA gene trees. The work presented here represents the first comprehensive, genome- scale taxonomic analysis of the entirety of the order ‘Enterobacteriales’.