Realizing Microbial Evolution
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Global Transposon Mutagenesis and a Minimal Mycoplasma Genome
R EPORTS thermore, while immunosubunit precursors- stained positive, whereas no staining of PA28Ϫ/Ϫ purchased from PharMingen (San Diego, CA). 25D1.16 containing 15S proteasome assembly inter- cells was observed. (10) and a PA28a-specific antiserum (2) were kindly 11. S. C. Jameson, F. R. Carbone, M. J. Bevan, J. Exp. Med. provided by A. Porgador and M. Rechsteiner, respective- mediates were detected in wild-type cells, 177, 1541 (1993). ly. Metabolic labeling, immunoprecipitation, immuno- under identical conditions 15S complexes 12. T. Preckel and Y. Yang, unpublished results. blotting, and sodium dodecyl sulfateÐpolyacrylamide were hardly detectable in PA28Ϫ/Ϫ cells (Fig. 13. M. Ho, Cytomegalovirus: Biology and Infection (Ple- gel electrophoresis (SDS-PAGE) were performed as de- 4B). Notably, PA28 was found to be associ- num, New York, ed. 2, 1991). scribed (3). 14. P. Borrow and M. B. A. Oldstone, in Viral Pathogenesis, 19. T. Flad et al., Cancer Res. 58, 5803 (1998). ated with the immunosubunit precursor-con- N. Nathanson, Ed. (Lippincott-Raven, Philadelphia, 20. M. W. Moore, F. R. Carbone, M. J. Bevan, Cell 54, 777 taining 15S complexes, suggesting that PA28 1997), pp. 593Ð627; M. J. Buchmeier and A. J. Zajac, (1988). is required for the incorporation of immuno- in Persistent Virus Infections, R. Ahmed and I. Chen, 21. A. Vitiello et al., Eur. J. Immunol. 27, 671 (1997). Eds. ( Wiley, New York, 1999), pp. 575Ð605. subunits into proteasomes. 22. H. Bluthmann et al., Nature 334, 156 (1988). 15. G. Niedermann et al., J. Exp. Med. 186, 209 (1997); 23. W. Jacoby, P. V. Cammarata, S. -

The Transcriptional Landscape of a Rewritten Bacterial Genome Reveals Control Elements and Genome Design Principles ✉ ✉ Mariëlle J
ARTICLE https://doi.org/10.1038/s41467-021-23362-y OPEN The transcriptional landscape of a rewritten bacterial genome reveals control elements and genome design principles ✉ ✉ Mariëlle J. F. M. van Kooten 1 , Clio A. Scheidegger1, Matthias Christen1 & Beat Christen 1 Sequence rewriting enables low-cost genome synthesis and the design of biological systems with orthogonal genetic codes. The error-free, robust rewriting of nucleotide sequences can 1234567890():,; be achieved with a complete annotation of gene regulatory elements. Here, we compare transcription in Caulobacter crescentus to transcription from plasmid-borne segments of the synthesized genome of C. ethensis 2.0. This rewritten derivative contains an extensive amount of supposedly neutral mutations, including 123’562 synonymous codon changes. The tran- scriptional landscape refines 60 promoter annotations, exposes 18 termination elements and links extensive transcription throughout the synthesized genome to the unintentional intro- duction of sigma factor binding motifs. We reveal translational regulation for 20 CDS and uncover an essential translational regulatory element for the expression of ribosomal protein RplS. The annotation of gene regulatory elements allowed us to formulate design principles that improve design schemes for synthesized DNA, en route to a bright future of iteration- free programming of biological systems. 1 Institute of Molecular Systems Biology, Department of Biology, Eidgenössische Technische Hochschule Zürich, Zürich, Switzerland. ✉ email: [email protected]; [email protected] NATURE COMMUNICATIONS | (2021) 12:3053 | https://doi.org/10.1038/s41467-021-23362-y | www.nature.com/naturecommunications 1 ARTICLE NATURE COMMUNICATIONS | https://doi.org/10.1038/s41467-021-23362-y e can program biological systems with DNA that is In previous work, we synthesized and assembled the rewritten Wbased on native nucleotide sequences. -

Horizontal Gene Flow Into Geobacillus Is Constrained by the Chromosomal Organization of Growth and Sporulation
bioRxiv preprint doi: https://doi.org/10.1101/381442; this version posted August 2, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Horizontal gene flow into Geobacillus is constrained by the chromosomal organization of growth and sporulation Alexander Esin1,2, Tom Ellis3,4, Tobias Warnecke1,2* 1Molecular Systems Group, Medical Research Council London Institute of Medical Sciences, London, United Kingdom 2Institute of Clinical Sciences, Faculty of Medicine, Imperial College London, London, United Kingdom 3Imperial College Centre for Synthetic Biology, Imperial College London, London, United Kingdom 4Department of Bioengineering, Imperial College London, London, United Kingdom *corresponding author ([email protected]) 1 bioRxiv preprint doi: https://doi.org/10.1101/381442; this version posted August 2, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Abstract Horizontal gene transfer (HGT) in bacteria occurs in the context of adaptive genome architecture. As a consequence, some chromosomal neighbourhoods are likely more permissive to HGT than others. Here, we investigate the chromosomal topology of horizontal gene flow into a clade of Bacillaceae that includes Geobacillus spp. Reconstructing HGT patterns using a phylogenetic approach coupled to model-based reconciliation, we discover three large contiguous chromosomal zones of HGT enrichment. -

Section 4. Guidance Document on Horizontal Gene Transfer Between Bacteria
306 - PART 2. DOCUMENTS ON MICRO-ORGANISMS Section 4. Guidance document on horizontal gene transfer between bacteria 1. Introduction Horizontal gene transfer (HGT) 1 refers to the stable transfer of genetic material from one organism to another without reproduction. The significance of horizontal gene transfer was first recognised when evidence was found for ‘infectious heredity’ of multiple antibiotic resistance to pathogens (Watanabe, 1963). The assumed importance of HGT has changed several times (Doolittle et al., 2003) but there is general agreement now that HGT is a major, if not the dominant, force in bacterial evolution. Massive gene exchanges in completely sequenced genomes were discovered by deviant composition, anomalous phylogenetic distribution, great similarity of genes from distantly related species, and incongruent phylogenetic trees (Ochman et al., 2000; Koonin et al., 2001; Jain et al., 2002; Doolittle et al., 2003; Kurland et al., 2003; Philippe and Douady, 2003). There is also much evidence now for HGT by mobile genetic elements (MGEs) being an ongoing process that plays a primary role in the ecological adaptation of prokaryotes. Well documented is the example of the dissemination of antibiotic resistance genes by HGT that allowed bacterial populations to rapidly adapt to a strong selective pressure by agronomically and medically used antibiotics (Tschäpe, 1994; Witte, 1998; Mazel and Davies, 1999). MGEs shape bacterial genomes, promote intra-species variability and distribute genes between distantly related bacterial genera. Horizontal gene transfer (HGT) between bacteria is driven by three major processes: transformation (the uptake of free DNA), transduction (gene transfer mediated by bacteriophages) and conjugation (gene transfer by means of plasmids or conjugative and integrated elements). -
Bacterial Genetics a Tiny Alternative
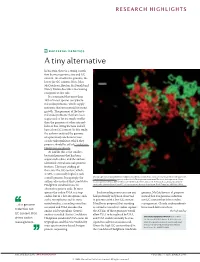
RESEARCH HIGHLIGHTS BACTERIAL GENETICS A tiny alternative In bacteria, there is a strong correla- tion between genome size and GC content: the smaller the genome, the lower the GC content. Now, John McCutcheon, Bradon McDonald and Nancy Moran describe a fascinating exception to this rule. It is estimated that more than 10% of insect species carry bacte- rial endosymbionts, which supply nutrients that are essential for insect growth. The genomes of the bacte- rial endosymbionts that have been sequenced so far are much smaller than the genomes of other intracel- lular or free-living bacteria and all have a low GC content. In this study, the authors analysed the genome of a previously uncharacterized cicada endosymbiont, which they propose should be called Candidatus Hodgkinia cicadicola. At 144 kb, this is the smallest bacterial genome that has been sequenced to date, and the authors identified several unusual genomic features. The most striking of these was the GC content, which, at 58%, is unusually high for such Micrograph showing Candidatus Hodgkinia cicadicola (red) in close association with another endosymbiont, a small genome. Intriguingly, the Candidatus Sulcia muelleri (green) in the cicada Diceroprocta semicincta. The scale bar represents 10 µm. authors also noticed that Candidatus Image reproduced from McCutcheon, J. P., McDonald, B. R. & Moran, N. P. Origin of an alternative genetic Hodgkinia cicadicola uses an code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 5, e1000565 (2009). alternative genetic code. In most species the codon UGA is a stop Such recoding events are rare and genome, McCutcheon et al. -

Synthetic Biology Projects in Vitro
Downloaded from genome.cshlp.org on September 25, 2021 - Published by Cold Spring Harbor Laboratory Press Review Synthetic biology projects in vitro Anthony C. Forster1,3 and George M. Church2,3 1Department of Pharmacology and Vanderbilt Institute of Chemical Biology, Vanderbilt University Medical Center, Nashville, Tennessee 37232, USA; 2Department of Genetics, Harvard Medical School, Boston, Massachusetts 02115, USA Advances in the in vitro synthesis and evolution of DNA, RNA, and polypeptides are accelerating the construction of biopolymers, pathways, and organisms with novel functions. Known functions are being integrated and debugged with the aim of synthesizing life-like systems. The goals are knowledge, tools, smart materials, and therapies. Synthetic biology projects (SBPs) an action plan to regulate world suppliers of DNA synthesizers, DNA precursors, and oligos (Church 2004). Ethical and safety The basic elements of chemistry and biology are few, but the issues have been, and must continue to be, regulated (Cho et al. synthetic combinations are unlimited and awe inspiring. The 1999). first international conference on synthetic biology charted its Several groups have proposed to create bacteria with chro- goals as understanding and utilizing life’s diverse solutions to mosomes synthesized entirely from synthetic oligos. This might process information, materials, and energy (Silver and Way 2004) be done stepwise (Posfai et al. 2006) or by inactivating the en- (http://syntheticbiology.org). As a bonus, genetic systems are dogenous bacterial chromosome and then somehow transform- biocompatible, renewable, and can be optimized by Darwinian ing and rebooting the bacterium with an entire in vitro- selections. SBPs entail the complex manipulation of replicating synthesized genome. -

Assessment of the Bimodality in the Distribution of Bacterial Genome Sizes
The ISME Journal (2017) 11, 821–824 © 2017 International Society for Microbial Ecology All rights reserved 1751-7362/17 www.nature.com/ismej SHORT COMMUNICATION Assessment of the bimodality in the distribution of bacterial genome sizes Hyun S Gweon, Mark J Bailey and Daniel S Read Centre for Ecology & Hydrology, Wallingford, UK Bacterial genome sizes have previously been shown to exhibit a bimodal distribution. This phenomenon has prompted discussion regarding the evolutionary forces driving genome size in bacteria and its ecological significance. We investigated the level of inherent redundancy in the public database and the effect it has on the shape of the apparent bimodal distribution. Our study reveals that there is a significant bias in the genome sequencing efforts towards a certain group of species, and that correcting the bias using species nomenclature and clustering of the 16S rRNA gene, results in a unimodal rather than the previously published bimodal distribution. The true genome size distribution and its wider ecological implications will soon emerge as we are currently witnessing rapid growth in the number of sequenced genomes from diverse environmental niches across a range of habitats at an unprecedented rate. The ISME Journal (2017) 11, 821–824; doi:10.1038/ismej.2016.142; published online 11 November 2016 Significant progress has been made in understanding Wolf in 2008, where it was reported that bacterial interactions between ecology and genome evolution genome sizes show a bimodal distribution (Koonin in prokaryotes. -

Development of an Artificial Cell, from Self- INAUGURAL ARTICLE Organization to Computation and Self-Reproduction
Development of an artificial cell, from self- INAUGURAL ARTICLE organization to computation and self-reproduction Vincent Noireauxa, Yusuke T. Maedab, and Albert Libchaberb,1 aUniversity of Minnesota, 116 Church Street SE, Minneapolis, MN 55455; and bThe Rockefeller University, 1230 York Avenue, New York, NY 10021 This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected in 2007. Contributed by Albert Libchaber, November 22, 2010 (sent for review October 13, 2010) This article describes the state and the development of an artificial the now famous “Omnis cellula e cellula.” Not only is life com- cell project. We discuss the experimental constraints to synthesize posed of cells, but also the most remarkable observation is that a the most elementary cell-sized compartment that can self-reproduce cell originates from a cell and cannot grow in situ. In 1665, Hooke using synthetic genetic information. The original idea was to program made the first observation of cellular organization in cork mate- a phospholipid vesicle with DNA. Based on this idea, it was shown rial (8) (Fig. 1). He also coined the word “cell.” Schleiden later that in vitro gene expression could be carried out inside cell-sized developed a more systematic study (9). The cell model was finally synthetic vesicles. It was also shown that a couple of genes could fully presented by Schwann in 1839 (10). This cellular quantiza- be expressed for a few days inside the vesicles once the exchanges tion was not a priori necessary. Golgi proposed that the branched of nutrients with the outside environment were adequately intro- axons form a continuous network along which the nervous input duced. -

Commentary the Minimal Cell Genome: ''On Being the Right Size”
Proc. Natl. Acad. Sci. USA Vol. 93, pp. 10004–10006, September 1996 Commentary The minimal cell genome: ‘‘On being the right size” Jack Maniloff* Department of Microbiology and Immunology, University of Rochester, Medical Center Box 672, Rochester, NY 14642 ‘‘On being the right size’’ is in quotation marks in the title these studies, it is important to remember that organisms with because it has been used twice before–in each case to discuss small genomes have arisen by two vastly different evolutionary the size of biological systems in terms of interest at that time. pathways, one ‘‘top down’’ and the other ‘‘bottom up.’’ J. B. S. Haldane (1) originally used this title in the 1920s, for ‘‘Top down’’ genomes evolved in organisms with increasing a paper describing the relationship between animal size and metabolic requirements. At each metabolic level, there can be constraints like gravity, surface tension, and food and oxygen a minimal genome: from photoautotrophs, able to grow in a consumption. In the 1970s, N. W. Pirie (2) (acknowledging medium of only CO2, light, and inorganic salts; to simple Haldane) used the title for a paper analyzing how factors like heterotrophs, for which growth requires a basic medium membrane properties, water structure, and the volume needed containing an organic carbon and energy source (like glucose) for ribosomes and other macromolecules set lower limits on and inorganic salts; to fastidious heterotrophs, for which cell size. Now, in the 1990s, the title is used to discuss efforts growth requires a complex medium, frequently containing to define the minimal genome content necessary and sufficient undefined components (like serum); to obligate intracellular for a living cell. -

Mechanisms Of, and Barriers To, Horizontal Gene Transfer Between Bacteria
FOCUS ON HORIZONTAL GENE TRANSFER MECHANISMS OF, AND BARRIERS TO, HORIZONTAL GENE TRANSFER BETWEEN BACTERIA Christopher M. Thomas* and Kaare M. Nielsen‡ Abstract | Bacteria evolve rapidly not only by mutation and rapid multiplication, but also by transfer of DNA, which can result in strains with beneficial mutations from more than one parent. Transformation involves the release of naked DNA followed by uptake and recombination. Homologous recombination and DNA-repair processes normally limit this to DNA from similar bacteria. However, if a gene moves onto a broad-host-range plasmid it might be able to spread without the need for recombination. There are barriers to both these processes but they reduce, rather than prevent, gene acquisition. The first evidence that horizontal gene transfer (HGT) informational genes of the central cellular machinery could occur was the recognition that virulence deter- such as DNA replication, transcription or translation minants could be transferred between pneumococci in tend not to spread rapidly, even if they confer anti biotic infected mice, a phenomenon that was later shown to resistance, compared for example to single-function- be mediated by the uptake of the genetic material DNA resistance determinants such as β-lactamases or in a process called transformation1. The subsequent aminoglycoside-modifying enzymes. However, the identification of gene transfer mediated by both plas- nature of the transfer mechanism can also determine the mids and viruses and the recognition of transposable organisms and genes that are most often involved. The elements provided the stepping stones to our current purpose of this review is to describe some of the mecha- picture of gene flux and the importance of mobile nisms that lead to horizontal gene acquisitions with a genetic elements2. -

Synthetic Biology As a Technoscience: the Case of Minimal Genomes and Essential Genes
Published in Studies in History and Philosophy of Science (epub ahead of print) Synthetic Biology as a Technoscience: The Case of Minimal Genomes and Essential Genes Massimiliano Simons, Ghent University [email protected] Department of Philosophy and Moral Sciences, Blandijnberg 2, BE-9000, Ghent Abstract This article examines how minimal genome research mobilizes philosophical concepts such as minimality and essentiality. Following a historical approach the article aims to uncover what function this terminology plays and which problems are raised by them. Specifically, four historical moments are examined, linked to the work of Harold Morowitz, Mitsuhiro Itaya, Eugene Koonin and Arcady Mushegian, and J. Craig Venter. What this survey shows is a historical shift away from historical questions about life or descriptive questions about specific organisms towards questions that explore biological possibilities: what are possible forms of minimal genomes, regardless of whether they exist(ed) in nature? Moreover, it highlights a fundamental ambiguity at work in minimal genome research between a universality claim and a standardization claim: does a minimal genome refer to the minimal gene set for any organism whatsoever? Or does it refer rather to a gene set that will provide stable, robust and predictable behaviour, suited for biotechnological applications? Two diagnoses are proposed for this ambiguity: a philosophical diagnosis of how minimal genome research either misunderstands the ontology of biological entities or philosophically misarticulates scientific practice. Secondly, a historical diagnosis that suggests that this ambiguity is part of a broader shift towards technoscience. Keywords: Minimal genome ; essential gene; Mycoplasma ; Craig Venter; synthetic biology ; technoscience Competing interests No competing interests to declare. -

Selection and Robustness in Bacterial Genome Evolution Seila Omer University of Connecticut, [email protected]
University of Connecticut OpenCommons@UConn Doctoral Dissertations University of Connecticut Graduate School 12-16-2016 Selection and Robustness in Bacterial Genome Evolution Seila Omer University Of Connecticut, [email protected] Follow this and additional works at: https://opencommons.uconn.edu/dissertations Recommended Citation Omer, Seila, "Selection and Robustness in Bacterial Genome Evolution" (2016). Doctoral Dissertations. 1317. https://opencommons.uconn.edu/dissertations/1317 Selection and Robustness in Bacterial Genome Evolution Seila Omer, Ph.D. University of Connecticut, 2016 The research presented in this thesis attempts to address research questions related to the role of natural selection in the evolution of bacterial genes not expressed for function and in building mutational tolerance to translational errors. Studies on evolution of protein coding DNA sequences have provided the evidence for a current paradigm in evolutionary biology: only functional genes are undergoing selection against the deleterious effects of allele variants (purifying selection). I provide evidence that similar footprints of selection can be detected in genes that are not normally expressed for function during the bacterial life cycle. Using simulations for DNA sequence evolution, I demonstrate statistically significant deviations from neutral evolution for the studied genes. I suggest that purifying selection affects both functional and non-functional genes. I propose this might be caused by the dominant toxic effects of low level translation of mutated products in bacteria, due to misfolding and misinteraction. Natural selection also acts to remove the effects of translational errors. Stop codon readthrough events are more likely to have major structural and functional effects than simple nucleotide changes. Recent research has shown that strength of selection experienced by protein-coding genes is positively correlated with the level of gene expression.