Studies of Mitochondrial DNA, Allozyme and Morphometric Variation in a House Mouse Hybrid Zone
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Spezzoni Pari O Inferiori a Sei Ore Primo Grado
Ministero dell’Istruzione, dell’Università e della Ricerca Ufficio Scolastico Regionale per la Lombardia Ufficio XX – Sondrio ANNO SCOLASTICO 2013/2014 Classi di concorso SPEZZONI PARI O INFERIORI A 6 ORE Scuola secondaria I grado AB77 - chitarra 6 ore SONDRIO “Torelli” AC77 - clarinetto 6 ore SONDRIO “Torelli” AG77 - flauto 6 ore PONTE IN VALTELLINA AJ77 - pianoforte 6 ore BORMIO “Anzi” 6 ore SONDRIO “Torelli” AM77 – violino 6 ore SONDRIO “Torelli” 2 ore BERBENNO DI VALTELLINA A028 - Arte e immagine 2 ore VALDISOTTO 2 ore VILLA DI CHIAVENNA 4 ore CAMPODOLCINO 4 ore DUBINO 4 ore GROSOTTO 6 ore LIVIGNO 2 ore PONTE IN VALTELLINA 2 ore TIRANO 2 ore BERBENNO DI VALTELLINA A030 – Scienze motorie e sportive 2 ore VILLA DI CHIAVENNA 4 ore CAMPODOLCINO 6 ore LIVIGNO 4 ore MORBEGNO 6 ore PONTE IN VALTELLINA 6 ore APRICA 2 ore VILLA DI CHIAVENNA A032 - Musica 2 ore CHIAVENNA “Garibaldi” 4 ore CAMPODOLCINO 2 ore DUBINO 6 ore LIVIGNO 4 ore MORBEGNO 6 ore PONTE IN VALTELLINA 2 ore SONDRIO “Sassi” 2 ore ALBOSAGGIA 6 ore APRICA 2 ore BERBENNO DI VALTELLINA A033 - Tecnologia 2 ore VALDISOTTO 2 ore VILLA DI CHIAVENNA 4 ore CAMPODOLCINO 6 ore DUBINO USR Lombardia – Ufficio XX – Ambito territoriale di Sondrio – Via Donegani, 5 - 23100 Sondrio Tel. +39 0342 54 11 11 – Email [email protected] 6 ore LIVIGNO 4 ore MORBEGNO 6 ore PONTE IN VALTELLINA 4 ore ALBOSAGGIA 4 ore TALAMONA 6 ore APRICA 4 ore VALDISOTTO A043 - Italiano, storia e geografia nella scuola 1 ora VALFURVA secondaria di I grado 6 ore DUBINO 2 ore GROSOTTO 3 ore PONTE IN VALTELLINA 2 ore SONDRIO “Piazzi” 4 ore TALAMONA 4 ore VILLA DI TIRANO 2 ore DELEBIO C.T.P. -
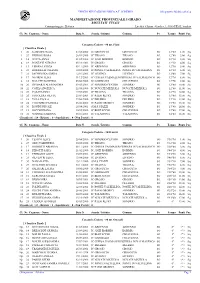
Classifiche Atletica 2014 Cadette Indiv
UFFICIO EDUCAZIONE FISICA A.T. SONDRIO Olimpyawin Modulo atletica MANIFESTAZIONE PROVINCIALE I GRADO RISULTATI FINALI Cronometraggio : Elettrico Località :Chiuro - Giudice A.:GGG FIDAL Sondrio Cl. Nr. Cognome - Nome Data N. Scuola / Istituto Comune Pv Tempo Punti Par. Categoria Cadette - 80 mt. Piani [ Classifica Finale ] 1 20 GAMMINO GAIA 07/08/2000 IC GROSOTTO GROSOTTO SO 12"00 1,00 Sq. 1 27 PEDROLI ROSA 26/03/2000 IC TIRANO TIRANO SO 12"00 2,00 Sq. 3 14 SOTTA ANNA 01/05/2000 IC ANZI BORMIO BORMIO SO 12"10 3,00 Sq. 4 19 RANZANI AURORA 05/10/2001 IC GROSIO GROSIO SO 12"30 4,00 Sq. 4 13 LIBERA LETIZIA 05/11/2000 IC ARDENNO ARDENNO SO 12"30 5,00 Sq. 6 23 BORINELLI CELESTE 10/08/2000 IC PONTE VALTELLINA PONTE IN VALTELLINA SO 12"40 6,00 Sq. 7 21 JACHEVSKA SOFIJA 12/01/2001 IC LIVIGNO LIVIGNO SO 12"60 7,00 Sq. 8 17 NEGRINI ELISA 31/12/2001 IC CHIESA VALMALENCOCHIESA IN VALMALENCO SO 12"70 8,00 Sq. 8 16 BALATTI MARTINA 05/06/2000 IC GARIBALDI CHIAVENNA SO 12"70 9,00 Sq. 10 26 DE MARZI ALESSANDRA 09/05/2001 IC SONDRIO CENTRO SONDRIO SO 12"80 10,00 Sq. 10 22 COPES ANGELICA 22/05/2000 IC NOVATE MEZZOLA NOVATE MEZZOLA SO 12"80 11,00 Sq. 12 29 CASATI ANNA 17/04/2001 IC TRAONA TRAONA SO 12"90 12,00 Sq. 13 25 FISTOLERA SILVIA 23/09/2001 IC PAESI RETICI SONDRIO SO 13"00 13,00 Sq. -

ESITO 1A PROVA OPERATORI QUALIFICATI 16-05-14
ELENCO DEGLI ISCRITTI AL CORSO PER OPERATORI CONTROLLO CINGHIALE, AMMESSI ALLA PROVA PRATICA NOMINATIVO DATA NASCITA RESIDENZA AGNELLI BASILIO 31/01/1960 CASPOGGIO AMBROSINI FABIO 26/02/1971 ARDENNO AMONINI LUCIANO 17/12/1960 CASTELLO DELL'ACQUA ANDREOLI OSVALDO 02/08/1959 BERBENNO DI VALT. ANDREOTTA GUGLIELMO 30/10/1961 VILLA DI TIRANO BAGIOLO MICHELE 16/11/1986 CHIESA IN VALMALENCO BAGIOTTI JOHNY 21/02/1970 POSTALESIO BARLASCINI MARIO 28/12/1959 TARTANO BASSI SIMONETTA 09/03/1967 ARDENNO BASSOLA GIANCARLO 10/08/1968 SONDRIO BERTOLINI FULVIO 19/06/1940 TALAMONA BONETTI DINO 18/03/1972 VALDISOTTO BRACCHI ALBERTO 20/03/1982 GROSOTTO BRAGA GIOVANNI 03/06/1969 TALAMONA BRICALLI GIULIANO 21/11/1949 CASPOGGIO CANTONI ALDO 19/09/1955 PIATEDA CAO GIOVANNI 20/04/1962 TIRANO CATTALINI FRANCO 30/06/1943 VILLA DI TIRANO CATTALINI PATRIZIA 24/09/1968 CHIURO CECINI INNOCENTE 10/09/1963 GROSOTTO CECINI TOMASO 03/10/1966 TOVO S.AGATA CERIBELLI ERNESTO 25/12/1950 SONDRIO CERIBELLI FRANCESCO 15/04/1988 SONDRIO COLOMBINI ANDREA 20/03/1976 VILLA DI TIRANO COMOLATTI LUIGI 04/09/1962 SONDRIO CONGIU SILVIO 28/11/1952 PONTE IN VALT. CONTRIO CIRILLO 16/02/1947 ALBOSAGGIA D'ALPAOS MAURIZIO 08/01/1958 SONDRIO DE FILIPPI FILIPPO 28/07/1948 TIRANO DE PAOLI LUCIANO WALTER 06/11/1948 TRESIVIO DEI CAS IVAN 16/07/1980 VALDISOTTO DEL PIANO GIANCARLO 08/06/1960 CHIURO DELLA MADDALENA GIANFRANCO 01/08/1955 MONTAGNA IN VALT. DELL'AVANZO GIANNI 19/09/1952 PONTE IN VALT. DERADA PIERGIACOMO 30/03/1963 VILLA DI TIRANO DIOLI ELIO 24/10/1960 CASPOGGIO FANETTI GIANFRANCO 10/12/1964 PRATA CAMPORTACCIO FERRARO DARIO 08/10/1960 DUBINO FORTINI VITTORIO 28/01/1944 ALBOSAGGIA ELENCO DEGLI ISCRITTI AL CORSO PER OPERATORI CONTROLLO CINGHIALE, AMMESSI ALLA PROVA PRATICA NOMINATIVO DATA NASCITA RESIDENZA FRANCESCHINA JESSICA 03/08/1979 BORMIO FRANCESCHINA JONATHAN 28/06/1978 VALDIDENTRO FRANZETTI MARIO 26/05/1959 BERBENNO DI VALT. -

Spezzoni I Grado
Ministero dell’Istruzione, dell’Università e della Ricerca ISTITUTO COMPRENSIVO SONDRIO “PAESI RETICI” Via DonLucchinetti, 3 – 23100 SONDRIO ANNO SCOLASTICO 2014/2015 Classi di concorso SPEZZONI PARI O INFERIORI A 6 ORE Scuola secondaria I grado AG77 - flauto 6 ore PONTE IN VALTELLINA AJ77 - pianoforte 6 ore BORMIO “Anzi 2 ore BERBENNO DI VALTELLINA A028 - Arte e immagine 2 ore VALFURVA 2 ore VILLA DI CHIAVENNA 2 ore CHIAVENNA “Garibaldi” 2 ore GORDONA 6 ore CAMPODOLCINO 4 ore CHIESA IN VALMALENCO 2 ore DELEBIO 2 ore DUBINO 4 ore GROSOTTO 6 ore LIVIGNO 6 ore MORBEGNO “Vanoni” 2 ore SAMOLACO 6 ore PONTE IN VALTELLINA 4 ore SONDRIO “Ligari” 6 ore ALBOSAGGIA 6 ore SONDRIO “Torelli” 2 ore SONDRIO “Piazzi” 2 ore TIRANO 2 ore TRAONA 2 ore BERBENNO DI VALTELLINA A030 – Scienze motorie e sportive 2 ore VILLA DI CHIAVENNA 2 ore CHIAVENNA “Garibaldi” 2 ore GORDONA 6 ore CAMPODOLCINO 4 ore DUBINO 6 ore LIVIGNO 2 ore SAMOLACO 6 ore PONTE IN VALTELLINA 2 ore SONDRIO “Sassi” 6 ore APRICA 6 ore BOR MIO A032 - Musica 2 ore VALFURVA 2 ore VILLA DI CHIAVENNA 2 ore CHIAVENNA “Garibaldi” 6 ore CAMPODOLCINO 6 ore LIVIGNO 4 ore NOVATE MEZZOLA 2 ore SONDRIO “Sassi” 4 ore ALBOSAGGIA 6 ore APRICA 2 ore VAL FURVA A033 - Tecnologia 2 ore CHIAVENNA “Bertacchi” 4 ore VILLA DI CHIAVENNA 2 ore CHIAVENNA “Garibaldi” 6 ore CAMPODOLCINO 6 ore LIVIGNO 6 ore MORBEGNO “Damiani” 2 ore SAMOLACO 6 ore PONTE IN VALTELLINA 6 ore APRICA 6 ore VALDI DENTRO A043 - Italiano, storia e geografia nella scuola 4 ore VALFURVA secondaria di I grado 2 ore VILLA DI CHIAVENNA 6 ore GORDONA 2 ore GROSOTTO 2 ore SAMOLACO 3 ore PONTE IN VALTELLINA 6 ore TEGLIO 4 ore VILLA DI TIRANO 2 ore DELEBIO C.T.P. -

Publication of the Amended Single Document Following the Approval Of
22.6.2020 EN Offi cial Jour nal of the European Union C 208/13 Publication of the amended single document following the approval of a minor amendment pursuant to the second subparagraph of Article 53(2) of Regulation (EU) No 1151/2012 (2020/C 208/06) The European Commission has approved this minor amendment in accordance with the third subparagraph of Article 6(2) of Commission Delegated Regulation (EU) No 664/2014 (1). The application for approval of this minor amendment can be consulted in the Commission’s eAmbrosia database. SINGLE DOCUMENT ‘MELA DI VALTELLINA’ EU No: PGI-IT-0574-AM01 – 17.2.2020 PDO ( ) PGI (X) 1. Name(s) ‘Mela di Valtellina’ 2. Member State or third country Italy 3. Description of the agricultural product or foodstuff 3.1. Type of product Class 1.6 – Fruit, vegetables and cereals, fresh or processed. 3.2. Description of product to which the name in (1) applies The following varieties are used for the production of ‘Mela di Valtellina’: ‘Red Delicious’ – ‘Golden Delicious’ – ‘Gala’. When released for consumption, they have the following characteristics: Red Delicious: thick, non-waxy epicarp of a brilliant, intense red colour, with the dominant colour covering more than 80 % of the surface, smooth, with no russeting or greasiness, resistant to handling. Oblong truncated cone shape with five lobes and a pentagonal equatorial plane. Minimum diameter of 65 mm. Minimum sugar content of more than 10° Brix. Flesh: white with apple aroma of medium intensity. Intense honey, jasmine and apricot aromas. Very crunchy and juicy. Dominant sweet flavour with appreciable acidity and aroma of medium intensity. -

Piano Territoriale Regionale D'area Media E Alta
Piano Territoriale Regionale d’Area Media e Alta Valtellina PIANO TERRITORIALE REGIONALE D’AREA MEDIA E ALTA VALTELLINA Documento di Piano Relazione D.G. Territorio Urbanistica e Difesa del Suolo - U.O. Programmazione Territoriale e Urbanistica Struttura Urbanistica e Progetti per il Territorio 30 luglio 2013 Documento di Piano – Relazione aggiornamento 2019 1 Piano Territoriale Regionale d’Area Media e Alta Valtellina Elaborazione del Piano Territoriale Regionale d’Area Provincia di Sondrio Consorzio Parco Nazionale dello Stelvio CCIAA di Sondrio Ha collaborato Documento di Piano – Relazione aggiornamento 2019 2 Piano Territoriale Regionale d’Area Media e Alta Valtellina Provincia di Sondrio Evaristo Pini, Roberto Nella Regione Lombardia D.G. Territorio e Urbanistica - U.O. Programmazione e Pianficazione Territoriale - Struttura Progetti per il Territorio Maurizio Federici, Antonio Lampugnani, Chiara Penco ERSAF Coordinamento scientifico e metodologico: Dario Kian, Giovanna Fossa Gruppo di lavoro: Andrea Bigatti, Daniela Masotti, Filomena Pomilio, Lisa Garbellini, Luca Tepsich, Martina Nessi, Mattia Cattaneo, Simona Muscarino, Tomaso Pompili, Paolo Trivellato, Gabriele Borsani, Andreina Degli Esposti, Laura Sommaruga, Fabio Bonelli, Giancarlo Graci, Filippo Manfredi. Documento di Piano – Relazione aggiornamento 2019 3 Schema generale di struttura del Piano Territoriale d’Area Media e Alta Valtellina Documento di Piano Relazione Tavole di analisi e interpretazione Tavole delle scelte di piano Allegato I "Quadro programmatico Allegato -

Cognome E Nome Bonus Comune Di Residenza
GRADUATORIA PROVVISORIA "BONUS TRASPORTI " 2018-2019 - PER STUDENTI UNVERSITARI DELLA PROVINCIA DI SONDRIO COGNOME E NOME BONUS COMUNE DI RESIDENZA CIRELLI SARA 300 TALAMONA BAUDINI MONICA 300 ARDENNO CARTOLANO ALESSANDRO 300 DELEBIO MAMBRETTI MARIO 300 DELEBIO CASPANI LUCA 300 MAZZO DI VALTELLINA DE BERNARDI ROBERTO 300 MORBEGNO RUINA ISABELLA 300 SONDRIO SCERESINI BENEDETTA 300 SONDRIO SONGINI ELISABETTA 300 VALMASINO MEVIO CRISTIAN 300 CASTIONE ANDEVENNO TOME’ SIMONE 300 PONTE IN VALTELLINA ARMANINI ALESSIA 300 COLORINA MONTAGNA IN PIASINI BEATRICE 300 VALTELLINA GOBBI VERONICA 300 PIANTEDO FLORIT GIADA 300 SONDALO BARONA FRANCESCO 300 DUBINO CORNAGGIA RANIERI DOMENICO 300 COSIO VALTELLINO FASCENDINI MASSIMO 300 ARDENNO AGUTOLI SIMONE 300 TIRANO CRAPELLA FEDERICO 300 POGGIRIDENTI SPEZIALE CHIARA 300 MORBEGNO ANGIOLETTI CAMILLA 300 SONDRIO ANGIOLETTI MATTEO 300 SONDRIO BERTOLINI ALICE 300 MORBEGNO BERTONI LUCA 300 VILLA DI TIRANO NANA DANIELA 300 CHIESA IN VALMALENCO FOGLIATTI MARTINA 400 MORBEGNO BRICALLI TANIA 300 BERBENNO DI VAL. SPINI SARA 300 CHIAVENNA GARZELLI CRISTINA 300 MANTELLO VIVIANI MARTINA 300 VALDIDENTRO TOGNINI CHIARA 300 CASTIONE ANDEVENNO GOSATTI BEATRICE 300 SONDRIO CANEVA MAURO 400 TALAMONA DAL POZZO VIVIANA 300 SONDALO DEL FANTE DELIA 300 SAMOLACO CAPELLI JULIAN ALBERT 300 TIRANO ZIVERI SARA 300 DELEBIO ZUCCHI GIULIA 400 BERBENNO DI VALT. PREVOSTINI RACHELE 300 SONDALO CORNAGGIA ALESSIA 300 MORBEGNO MAZZINA FEDERICA 300 SAMOLACO GUSMEROLI ELENA 300 ARDENNO BONDANZA MATILDE LIDIA 300 ARDENNO TENTORI ELEONORA 300 TIRANO MAURO LORENZO 300 BIANZONE ANTONELLI FRANCESCO 300 MANTELLO VOLA MATTEO 300 TALAMONA ROSSATTI MARTINA 300 CASTELLO DELL'ACQUA AGUTOLI CHIARA 300 TIRANO ROMANI AURORA 300 BORMIO DE PAOLI ANDREA 300 BERBENNO DI VALT. FLEMATTI LISA 400 LANZADA BELLESINI CAMILLA 300 TIRANO INNOCENTI MARCO 300 ARDENNO BENVENUTI DAVIDE 300 SONDRIO RAMPELLINI MATTIA 300 MELLO SOSIO ALEX 300 VALFURVA BRADANINI MONICA 300 BORMIO VENTURINI SIMONE 300 PIATEDA VENINI LAURA 300 SONDRIO MELONI ALBERTO 300 BERBENNO DI VAL. -

TIRANO L'aquila Sul Castello
TIRANO l’aquila sul castello testo e grafica di Franco Visintin consulenza storica di Bruno Ciapponi Landi I Quaderni del gioved ì Associazione Culturale Valtellinesi a Milano Almeno due sono gli stemmi che la comunità tiranese si è data nei molti secoli della sua esistenza. aquila sovrastante un castello : antico stemma della famiglia Venosta, (ramo Vervio) feudataria del vescovo di Como e dell’ Imperatore. (chiave di volta di un portale di Vervio) da Foppoli San Martino, patrono di Tirano, dona al povero metà del suo mantello tagliandolo con la spada. (bassorilievo sopra il portale principale della Basilica della Madonna di Tirano) Due stemmi guerrieri dunque. Per narrare la storia della comunità tiranese ci si è ispirati quindi all’opuscolo “Tirano, città militare” curato da Eliana e Nemo Canetta per il Museo Etnografico Tiranese. Non può comunque essere dimenticata la storica opera di Don Egidio Pedrotti “Le fortificazioni di Tirano” (Giuffrè, 1960), pietra miliare su tale tema, alla quale si è fatto ampio riferimento. > “Tirano è detta castellum fin dal 1073. Se non murata doveva essere difesa da qualche opera militare, una rocca per lo meno, sin d’allora ” (Pedrotti). Forse per questo il Comune di Tirano ha adottato, già dal 1861, come stemma quello della famiglia Venosta che vede la nera aquila imperiale ad ali spiegate sul suo castello. Lo stemma qui raffigurato è a Tirano in Palazzo Salis e risale anch’esso al 1861. I Venosta di Match, provenienti dalla valle omonima (in latino Mazia ), laterale della Val Venosta, si insediarono a Mazzo intorno al XI sec. dopo aver ottenuto in feudo dal vescovo di Como i territori dell’an- tica pieve. -

Studies of Mitochondrial DNA, Allozyme and Morphometric Variation in a House Mouse Hybrid Zone
Genet. Res., Camb. (2002), 80, pp. 117–129. With 3 figures. # 2002 Cambridge University Press 117 DOI: 10.1017\S0016672302005773 Printed in the United Kingdom Studies of mitochondrial DNA, allozyme and morphometric variation in a house mouse hybrid zone HEIDI C. HAUFFE*, STELLA FRAGUEDAKIS-TSOLIS†, PATRICIA M. MIROL‡ JEREMY B. SEARLE Department of Biology, Uniersity of York, PO Box 373, York YO10 5YW, UK (Receied 17 April 2001 and in reised form 1 May 2002) Summary An unusual chromosomal hybrid zone of the house mouse, Mus musculus domesticus, exists in Upper Valtellina, Northern Italy, consisting of four Robertsonian (Rb) races and the standard (all- # acrocentric, or 2n l 40) race, all hybridizing freely within 10km. The hybrid zone in Valtellina provides an excellent opportunity to study the role of Rb fusions in reproductive isolation and speciation. This hybrid zone has already been well studied for the distribution of Rb fusions and the fertility of hybrids, but in order to understand the dynamics of the zone, a basic understanding of the origin and genetic similarity of the chromosomal races is necessary. This paper presents the results of three different methods of measuring genetic differentiation: multivariate analysis of morphological traits and analyses of allozyme variation and mitochondrial DNA sequences. The standard race is clearly distinguishable from the three Rb races by all three methods, but the Rb races are not distinguishable from one another. This provides strong evidence for our previous suggestions that the well-established Rb races in Valtellina are closely related, and that the standard race was introduced into the valley more recently from a distant source. -

Rationalisation of the Electricity Grid in the Valtellina Area: Agreement Reached Between Terna and Local Authorities for the Stretch Between Grosio and Tirano
RATIONALISATION OF THE ELECTRICITY GRID IN THE VALTELLINA AREA: AGREEMENT REACHED BETWEEN TERNA AND LOCAL AUTHORITIES FOR THE STRETCH BETWEEN GROSIO AND TIRANO The agreement plans 10 km of new underground power lines in the municipalities of Grosio, Grosotto and Mazzo di Valtellina and demolition of 29 km of existing overhead lines between Grosio and Tirano Rome, 7 May 2020 - After a year of work, and numerous meetings and discussions between Terna, the Province of Sondrio, the Mountain Community of Tirano and the Municipalities involved, today saw a first important milestone for that marks an initial phase of dismantling around 29 km of old power lines in the area between Grosio and Tirano, following creation of an underground connection running around 10 km through the municipalities of Grosio, Grosotto and Mazzo di Valtellina. The agreement between Terna and local authorities was reached in the context of the technical forum coordinated and organised by the Province of Sondrio, which took place via video conference to overcome restrictions imposed by the current Covid-19 emergency. Terna will soon launch further consultation with the municipalities involved to evaluate aspects for possible optimisation of the project and proceed with the authorisation process via the competent bodies. Dialogue has also begun with local areas for other stretches of the works. Phase A of this project for rationalisation of high-voltage electricity lines in the areas of Valtellina, Valchiavenna and Valcamonica, completed in 2017, has permitted the demolition of over 300 km of traditional power lines and creation of 180 km of underground lines to date. -

Elezioni U.P.S. 2020
ELEZIONI U.P.S. 2020 ELENCO ALFABETICO PESCATORI RESIDENTI IN PROVINCIA DI SONDRIO - VOTANTI/ELEGGIBILI Abela Sebastiano Sondrio Acquistapace Abele Piantedo Acquistapace Adriano Morbegno Acquistapace Andrea Piantedo Acquistapace Armando Piantedo Acquistapace Elia Samolaco Acquistapace Eraldo Piantedo Acquistapace Ezio Piantedo Acquistapace Marco Morbegno Acquistapace Marino Cosio Valtellino Acquistapace Maurizio Campodolcino Acquistapace Maurizio Piantedo Acquistapace Paolo Delebio Acquistapace Stefano Cosio Valtellino Acquistapace Stefano Delebio Acquistapace Thomas Cosio Valtellino Agnelli Andrea Caspoggio Agosti Gabriele Samolaco Agostinelli Franco Villa Di Tirano Aili Maurizio Berbenno Di Valtellina Aili Stefano Sondrio Albareda Ivan Chiesa In Valmalenco Albareda Mattia Chiesa In Valmalenco Albertazzi Rino Postalesio Alberti Edoardo Rasura Alberti Ottavio Ardenno ALBINI NICOLA Delebio ALBINIANO Luca Campodolcino Alessi Giuseppe Aprica Aloisio Paolo Samolaco Ambrosetti Remo Morbegno Ambrosetti Rocco Cosio Valtellino Ambrosini Attilio Aprica AMBROSINI CRISTIAN Dubino ambrosini daniele Cercino Ambrosini Fabio Ardenno Ambrosini Filippo Dubino Ambrosini Giuliano Delebio Ambrosini Lina Sondrio Ambrosini Valerio Dubino Ambrosini Valter Cercino Ambrosioni Luca Tresivio Amonini Antonello Castello Dell'acqua Amorini G.carlo Talamona Ancona Samuele Livigno Andreola Mattia Sondrio ANDREOLI ANDREA Mese Andreoli Giuliano Samolaco Andreoli Luca Tresivio ANDREOLI STEFANO Samolaco Andreotta Arnaldo Villa Di Tirano andreu caparros tamara Grosio Angelini -

Inventario a Cura Di C
Archivio storico del Comune di Talamona (Sondrio): sezione di Antico Regime Estremi cronologici: 1527 - 1801 Consistenza cronologica: 1398 - 1801 Introduzione e regesti a cura di Rita Pezzola Talamona, gennaio 2010 Abbreviazioni archivistiche utilizzate ACTlm Archivio del Comune di Talamona APTlm Archivio della Parrocchia di Talamona ASDCo Archivio storico della Diocesi di Como ASMi Archivio di Stato di Milano ASSo Archivio di Stato di Sondrio In copertina: Si tratta di un capolettera, speciale e ricercato. È la “T” del nome “Tognolus”, di un Tognolo Poletti abitante sul monte di Talamona. La vergò nel 1507 un notaio della Comunità di Talamona, attivissimo all’inizio del Cinquecento: Donato Camozzi, figlio di Guarisco, abitante nel borgo. Oggi questo capolettera può essere ripreso per indicare l’iniziale di Talamona. Ecco: un volto, un antico uomo della montagna, rivolto all’indietro, al passato; ma gli occhi - interpellanti - sono ben aperti sul presente. E dal copricapo, ricercato e adorno, sboccia un fiore offerto all’avvenire. Questa lettura simbolica - benché fuori delle intenzioni del notaio Camozzi - è fascinosa: per un rilancio dell’archivio e della storia istituzionale che vi è conservata e, nello stesso tempo, per il messaggio comunicativo non privo di echi poetici, che possono risuonare nell’animo di tutte le persone della comunità talamonese e ravvivare un senso di appartenenza. ASSo, Registri degli estimi, Talamona, 1, c. 117v. ARCHIVIO STORICO DEL COMUNE DI TALAMONA (SEZIONE DI ANTICO REGIME) Sommario Premessa III Introduzione V 1. Soggetti produttori:VI 1.1. Comune di Talamona: VI 1.2. Parrocchia di Talamona: XI 1.3. Luoghi pii (profili sintetici): XIII 1.3.1.