Analysis of Nectar from Low‐Volume Flowers
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
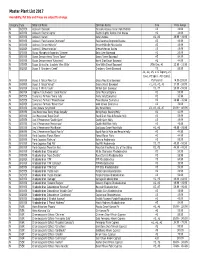
Master Plant List 2017.Xlsx
Master Plant List 2017 Availability, Pot Size and Prices are subject to change. Category Type Botanical Name Common Name Size Price Range N BREVER Azalea X 'Cascade' Cascade Azalea (Glenn Dale Hybrid) #3 49.99 N BREVER Azalea X 'Electric Lights' Electric Lights Double Pink Azalea #2 44.99 N BREVER Azalea X 'Karen' Karen Azalea #2, #3 39.99 - 49.99 N BREVER Azalea X 'Poukhanense Improved' Poukhanense Improved Azalea #3 49.99 N BREVER Azalea X 'Renee Michelle' Renee Michelle Pink Azalea #3 49.99 N BREVER Azalea X 'Stewartstonian' Stewartstonian Azalea #3 49.99 N BREVER Buxus Microphylla Japonica "Gregem' Baby Gem Boxwood #2 29.99 N BREVER Buxus Sempervirens 'Green Tower' Green Tower Boxwood #5 64.99 N BREVER Buxus Sempervirens 'Katerberg' North Star Dwarf Boxwood #2 44.99 N BREVER Buxus Sinica Var. Insularis 'Wee Willie' Wee Willie Dwarf Boxwood Little One, #1 13.99 - 21.99 N BREVER Buxus X 'Cranberry Creek' Cranberry Creek Boxwood #3 89.99 #1, #2, #5, #15 Topiary, #5 Cone, #5 Spiral, #10 Spiral, N BREVER Buxus X 'Green Mountain' Green Mountain Boxwood #5 Pyramid 14.99-299.99 N BREVER Buxus X 'Green Velvet' Green Velvet Boxwood #1, #2, #3, #5 17.99 - 59.99 N BREVER Buxus X 'Winter Gem' Winter Gem Boxwood #5, #7 59.99 - 99.99 N BREVER Daphne X Burkwoodii 'Carol Mackie' Carol Mackie Daphne #2 59.99 N BREVER Euonymus Fortunei 'Ivory Jade' Ivory Jade Euonymus #2 35.99 N BREVER Euonymus Fortunei 'Moonshadow' Moonshadow Euonymus #2 29.99 - 35.99 N BREVER Euonymus Fortunei 'Rosemrtwo' Gold Splash Euonymus #2 39.99 N BREVER Ilex Crenata 'Sky Pencil' -

2020-2021-Kube-Pak-Plug-Catalog
2 0 2 0 - 2 0 2 1 PLUGS & ROOTED CUTTINGS ECHINACEA ARTISAN SOFT ORANGE ANNUALS • PERENNIALS • CUT FLOWERS • VEGETABLES Echinacea Artisan Collection The NEW Artisan Collection™ elevates seed Echinacea to offer consistency of plant structure for uniform and highly branched, full plants. It also provides uniformity of flowering, so all plants are ready to sell at the same time. Plus, the benefit of a seed input provides flexibility for production so it fits better in your schedule. Height 16 to 28 in. (41 to 71 cm) Spread 10 to 25 in. (25 to 64 cm) Weeks to Finish 13 to 17 weeks Media Use well-drained, disease-free, soilless medium Artisan Red Ombre (pH 5.5 to 6.0, electrical conductivity [EC] 1.5-2.0 mS/cm) Temperature Days: 65-75°F (18-24°C) Nights: 60 to 65°F (16 to 18°C) Key Tips Too wet soil during overwinter production can result in Root Rot and uneven plant development. Short days are recommended to bulk the liners prior to long day conditions. Continue short day conditions until plants reach at least 7 fully mature leaves. After short day treatment, grow under long day conditions (natural or extended). Contact Kube-Pak today for Echinacea Artisan plugs for spring. See the full list of colors on page 48. Artisan Soft Orange Business Terms 2 Freight & Claims 3 Pricing Quick Reference 4 New Items 8 Drop List 10 Product List 2020-2021 13 Annuals from Seed 14 Annuals & Perennials from Cuttings 35 194 Route 526 Perennials from Seed 48 Allentown, NJ 08501 Cut Flowers 50 tel: (609) 259-3114 Vegetables & Fruits 51 fax: (609) 259-0487 Herbs 57 [email protected] Hardy Mums, Asters & Montauk Daisy 58 www.kubepak.com Grower’s Calendar 2020-2021 61 3% Early Order Discount Any orders for Spring 2021 placed by November 1, 2020 are eligible for our 3% Early Order Discount. -

Lamiales – Synoptical Classification Vers
Lamiales – Synoptical classification vers. 2.6.2 (in prog.) Updated: 12 April, 2016 A Synoptical Classification of the Lamiales Version 2.6.2 (This is a working document) Compiled by Richard Olmstead With the help of: D. Albach, P. Beardsley, D. Bedigian, B. Bremer, P. Cantino, J. Chau, J. L. Clark, B. Drew, P. Garnock- Jones, S. Grose (Heydler), R. Harley, H.-D. Ihlenfeldt, B. Li, L. Lohmann, S. Mathews, L. McDade, K. Müller, E. Norman, N. O’Leary, B. Oxelman, J. Reveal, R. Scotland, J. Smith, D. Tank, E. Tripp, S. Wagstaff, E. Wallander, A. Weber, A. Wolfe, A. Wortley, N. Young, M. Zjhra, and many others [estimated 25 families, 1041 genera, and ca. 21,878 species in Lamiales] The goal of this project is to produce a working infraordinal classification of the Lamiales to genus with information on distribution and species richness. All recognized taxa will be clades; adherence to Linnaean ranks is optional. Synonymy is very incomplete (comprehensive synonymy is not a goal of the project, but could be incorporated). Although I anticipate producing a publishable version of this classification at a future date, my near- term goal is to produce a web-accessible version, which will be available to the public and which will be updated regularly through input from systematists familiar with taxa within the Lamiales. For further information on the project and to provide information for future versions, please contact R. Olmstead via email at [email protected], or by regular mail at: Department of Biology, Box 355325, University of Washington, Seattle WA 98195, USA. -

A Phylogenetic Study of the Tribe Antirrhineae: Genome Duplications and Long-Distance Dispersals from the Old World to the New World 1
RESEARCH ARTICLE AMERICAN JOURNAL OF BOTANY A phylogenetic study of the tribe Antirrhineae: Genome duplications and long-distance dispersals from the Old World to the New World 1 Ezgi Ogutcen2 and Jana C. Vamosi PREMISE OF THE STUDY: Antirrhineae is a large tribe within Plantaginaceae. Mostly concentrated in the Mediterranean Basin, the tribe members are present both in the Old World and the New World. Current Antirrhineae phylogenies have diff erent views on taxonomic relationships, and they lack homogeneity in terms of geographic distribution and ploidy levels. This study aims to investigate the changes in the chromosome numbers along with dispersal routes as defi nitive characters identifying clades. METHODS: With the use of multiple DNA regions and taxon sampling enriched with de novo sequences, we provide an extensive phylogeny for Antir- rhineae. The reconstructed phylogeny was then used to investigate changes in ploidy levels and dispersal patterns in the tribe using ChromEvol and RASP, respectively. KEY RESULTS: Antirrhineae is a monophyletic group with six highly supported clades. ChromEvol analysis suggests the ancestral haploid chromosome number for the tribe is six, and that the tribe has experienced several duplications and gain events. The Mediterranean Basin was estimated to be the origin for the tribe with four long-distance dispersals from the Old World to the New World, three of which were associated with genome duplications. CONCLUSIONS: On an updated Antirrhineae phylogeny, we showed that the three out of four dispersals from the Old World to the New World were coupled with changes in ploidy levels. The observed patterns suggest that increases in ploidy levels may facilitate dispersing into new environments. -

Lianas and Climbing Plants of the Neotropics: Plantaginaceae
GUIDE TO THE GENERA OF LIANAS AND CLIMBING PLANTS IN THE NEOTROPICS PLANTAGINACEAE By Mark T. Strong (Jan 2021) A widely distributed family of primarily herbs, subshrubs or shrubs with about 100 genera and ca. 1,900 species worldwide. In the Neotropics, they are represented by about 45 genera and ca. 400 species. Five genera and 14 species are treated below as climbing plants. These occur in a diversity of habitats from desert scrub to montane cloud forests. Diagnostics: In vegetative condition, climbing Plantaginaceae have stems that are quadrangular or terete; leaves are opposite, alternate or sometimes verticillate, glabrous or glandular- pubescent, simple, and stipules are absent. In the order Lamiales, the quadrangular stems and simple opposite leaves of some Plantaginaceae Russelia syringifolia Schltdl. & Cham., photo by J. might be confused with Acanthaceae, Amith Gesneriaceae, Lamiaceae and Verbenaceae. Acanthaceae generally have ovaries with hook-like placental tissue and capsules with explosive dehiscence while in Gesneriaceae, ovaries are unilocular with parietal placentation. Lamiaceae and Verbenaceae have 2-ovulate ovaries and the fruit is a 4-parted schizocarp or dry indehiscent drupe. GENERAL CHARACTERS 1. STEMS. Quadrangular (sometimes winged) or terete in cross section, commonly solid, but hollow in some species of Russelia (e.g. R. campechiana Standl.; fig. 1a), xylem with deep phloem wedges in species of Russelia (e.g., R. contrerasii B.L. Turner; fig. 1b). Vessels narrow and commonly radially disposed (Metcalfe & Chalk, 1957). No visible exudates reported for the group. 2. PUBESCENCE. Glabrous or glandular-pubescent. 3. LEAVES. Alternate, opposite, verticillate or opposite proximally and alternate distally, the blades deltoid to cordiform, hastate or sagittate at base with palmate venation, or sometimes linear to lanceolate or ovate-lanceolate with pinnate venation, entire or with dentate margins, glabrous or glandular-pubescent; stipules absent. -

Diversity of Nectar Amino Acids in the Fritillaria(Liliaceae)
www.nature.com/scientificreports Corrected: Author Correction OPEN Diversity of nectar amino acids in the Fritillaria (Liliaceae) genus: ecological and evolutionary implications Katarzyna Roguz 1*, Andrzej Bajguz 2, Magdalena Chmur2, Agnieszka Gołębiewska2, Agata Roguz3 & Marcin Zych 1* Nectar is considered to be a primary food reward for most pollinators. It mostly contains sugars, but also has amino acids. The signifcance of the concentration and composition of amino acids in nectar is often less understood than that of its volume, sugar concentration and composition. However, there is a trend towards a broader approach in ecological research, which helps to understand nectar properties in an ecological context. The genus Fritillaria, exhibiting great diversity in fower morphology, nectar composition, and dominant pollinators, allows for the possibility to study some of the above. We studied the concentration and composition of amino acids in the nectar of 38 Fritillaria species attracting diferent groups of pollen vectors (bees, fies, passerines, and hummingbirds). The fowers of fritillaries produced nectar with a varying composition and concentration of amino acids. These diferences were mostly associated with the pollinator type. The nectar of passerine bird-pollinated species was rich in amino acids, whereas humming bird-pollinated produced low amino acid nectar. Contrary to previous reports nectar of the insect-pollinated species did not contain a higher amount of proline. Two non-protein amino acids, sarcosine and norvaline, were detected in the foral nectar for the frst time. Nectar is the most crucial foral reward for animal pollinators1. Primarily, it is a sugar solution composed of sucrose, fructose and glucose, in varying proportions. -

WUCOLS 2015 Plant List for So.Coastal Region.Xlsx
WUCOLS - South Coastal Region Type Botanical Name Common Name Water Use S Abelia chinensis Chinese abelia Unknown S Abelia floribunda Mexican abelia Moderate/Medium S Abelia mosanensis 'Fragrant Abelia' fragrant abelia Unknown S Abelia parvifolia (A. longituba) Schuman abelia Unknown Gc S Abelia x grandiflora and cvs. glossy abelia Moderate/Medium S Abeliophyllum distichum forsythia Unknown S Abelmoschus manihot (Hibiscus manihot) sunset muskmallow Unknown T Abies pinsapo Spanish fir Low T N Abies spp. (CA native and non-native) fir Moderate/Medium P N Abronia latifolia yellow sand verbena Very Low P N Abronia maritima sand verbena Very Low S N Abutilon palmeri Indian mallow Low S Abutilon pictum thompsonii variegated Chinese lantern Moderate/Medium S Abutilon vitifolium flowering maple Moderate/Medium S Abutilon x hybridum & cvs. flowering maple Moderate/Medium S T Acacia abyssinica Abyssinian acacia Inappropriate S Acacia aneura mulga Low S Acacia angustissima white ball acacia Unknown T Acacia baileyana Bailey acacia Low S T Acacia berlandieri guajillo Low S A Acacia boormanii Snowy River wattle Low T Acacia cognata (A.subporosa) bower wattle Moderate/Medium S T Acacia constricta whitethorn acacia Low S Acacia covenyi blue bush Low S T Acacia craspedocarpa leatherleaf acacia Low S Acacia cultriformis knife acacia Low T Acacia dealbata silver wattle Low T Acacia decurrens green wattle Low T Acacia erioloba camel thorn Low T Acacia farnesiana (See Vachellia farnesiana) Acacia farnesiana var. farnesiana (See T Vachellia farnesiana farnesiana) -

Surfinia® Heavenly Blue
Surfinia® Heavenly Blue Angel Earrings® (Fuchsia) Sundiascia® (Diascia) Desana® (Ipomoea) Sungelonia® (Angelonia) Ilumina® (Scoparia) Suntory® Chrysocephalum Lofos® (Lophospermum) (Chrysocephalum) Lofos® Compact (Lophospermum) Surdaisy® (Brachyscome) Marietta® (Bidens) Sunrdiva® (Scaevola) Million Bells Bouquet® (Calibrachoa) Surfinia® Bouquet (Petunia) Million Bells Mounding® (Calibrachoa) Surfinia® Mounding (Petunia) Million Bells Trailing® (Calibrachoa) Surfinia® Summer Double™ (Petunia) Moonlight EclipseTM (Petunia) Surfinia® Trailing (Petunia) Summer Splash® (Nierembergia) Tapien® (Verbena) Summer Wave® (Torenia) Temari ® Patio (Verbena) Summer Wave Bouquet® (Torenia) Temari ® Trailing (Verbena) SunbeliaTM (Lobelia) Violina® (Viola) 2 What’s New and Tried & True the Suntory Collection of annuals Delilah Onofrey starting on page 38. These include Flower Power Marketing Senetti, Sun Parasol, Sunvillea, for Suntory Collection Sunrosa, Crackling Fire and 440-522-1447 • [email protected] Princettia. Now you can find the entire Suntory portfolio of plants in e’re looking forward to another exciting year with five one place. Wnew series and nearly 20 new varieties that enhance • See all the exciting new varieties the Suntory brands you rely on. These new varieties are in one place on pages 6 and 7. improvements, new colors, superior performers and novelties • Choose plants that perform by that create interest. Each year, Suntory’s breeding teams sort using the recap of last summer’s through thousands of hybrids to select the very best. As new trials results on page 33. Which ones come in, others are phased out to offer manageable plants performed the best in your series with the very best performers. region? Throughout the catalog, you will see awards for specific varieties from the past three years. We are keeping You will notice some changes to the catalog this year: track to give you the most comprehensive view over time. -

Inter-Individual Nectar Chemistry Changes of Field Scabious, Knautia Arvensis
insects Article Inter-Individual Nectar Chemistry Changes of Field Scabious, Knautia arvensis Christine Venjakob 1,2, Sara Leonhardt 3 and Alexandra-Maria Klein 4,* 1 Ecosystem Functions, Institute of Ecology, Leuphana University of Lüneburg, Lüneburg, Universitätsallee 1, 21335 Lüneburg, Lower Saxony, Germany; [email protected] 2 Agroecology, Department of Crop Sciences, University of Göttingen, Grisebachstrasse 6, 37077 Göttingen, Lower Saxony, Germany 3 Biocenter—Am Hubland, Department of Animal Ecology and Tropical Biology, University of Würzburg, 97070 Würzburg, Bavaria, Germany; [email protected] 4 Chair of Nature Conservation and Landscape Ecology, Faculty of Environment and Natural Resources, University of Freiburg, Tennenbacher Str. 4, 79106 Freiburg, Baden-Württemberg, Germany * Correspondence: [email protected]; Tel.: +49-0761-203-67770 Received: 28 October 2019; Accepted: 15 January 2020; Published: 22 January 2020 Abstract: Nectar is crucial to maintain plant-pollinator mutualism. Nectar quality (nutritional composition) can vary strongly between individuals of the same plant species. The factors driving such inter-individual variation have however not been investigated closer. We investigated nectar quality of field scabious, Knautia arvensis in different grassland plant communities varying in species composition and richness to assess whether nectar quality can be affected by the surrounding plant community. We analyzed (with high performance liquid chromatography) the content of carbohydrates, overall amino acids, and essential amino acids. Amino acid and carbohydrate concentrations and proportions varied among plant individuals and with the surrounding plant community but were not related to the surrounding plant species richness. Total and individual carbohydrate concentrations were lowest, while proportions of the essential amino acids, valine, isoleucine, leucine (all phagostimulatory), and lysine were highest in plant species communities of the highest diversity. -

Annuals 2021
ANNUALS 2021 DISCLAIMER: The following list of plants offered this spring season was compiled using plants already in stock, listed on acknowledgments or from orders placed but not yet received. We make every effort to keep this list current and accurate before posting it online about April 15 of the current year. Before making the trip to Klein’s, it’s a good idea to give us a call to make sure that we actually have the sought after item in stock. Sometimes we don’t receive ordered items or sometimes items are substituted by our vendors at their discretion. Other times we experience a crop failure for whatever reason. And oftentimes we simply run out of product as the season progresses. In addition, pot or pack sizes are often changed at the last minute. Telephone: 608/244-5661 (or toll-free 888/244-5661) or email us at www.info@kleinsfloral.com. We’re sorry for any inconvenience. ACALYPHA PENDULA/HISPIDA (Chenille Plant): Bright red flowers are borne in long, pendent, tassel-like spikes. These tropical plants are best grown in light open shade with good air circulation. Keep moist. Allowing to dry too often will cause a rapid decline. Almost always grown in hanging baskets or containers. ‘Firetail’--Graceful, fuzzy, fiery-red cattail-like flowers appear at the tips of dark green foliage. Great, heat-loving plant. (5” pot and/or 10” hanging baskets) ACIDANTHERA, Gladiolus callianthus (Peacock Orchid): Cormous plant with linear leaves. In summer and in autumn, bears loose spikes of up to 10 hooded, funnel- shaped, strongly scented, pure white flowers, 2” across, each with a prominent purple- red mark in the throat. -

WUCOLS List S Abelia Chinensis Chinese Abelia M ? ? M / / Copyright © UC Regents, Davis Campus
Ba Bu G Gc P Pm S Su T V N Botanical Name Common Name 1 2 3 4 5 6 Symbol Vegetation Used in Type WUCOLS List S Abelia chinensis Chinese abelia M ? ? M / / Copyright © UC Regents, Davis campus. All rights reserved. bamboo Ba S Abelia floribunda Mexican abelia M ? M M / / S Abelia mosanensis 'Fragrant Abelia' fragrant abelia ? ? ? ? ? ? bulb Bu S Abelia parvifolia (A. longituba) Schuman abelia ? ? ? M ? ? grass G groundcover GC Gc S Abelia x grandiflora and cvs. glossy abelia M M M M M / perennial* P S Abeliophyllum distichum forsythia M M ? ? ? ? palm and cycad Pm S Abelmoschus manihot (Hibiscus manihot) sunset muskmallow ? ? ? L ? ? T Abies pinsapo Spanish fir L L L / / / shrub S succulent Su T N Abies spp. (CA native and non-native) fir M M M M / / P N Abronia latifolia yellow sand verbena VL VL VL / ? ? tree T P N Abronia maritima sand verbena VL VL VL / ? ? vine V California N native S N Abutilon palmeri Indian mallow L L L L M M S Abutilon pictum thompsonii variegated Chinese lantern M H M M ? ? Sunset WUCOLS CIMIS ET Representative Number climate 0 Region zones** Cities zones* S Abutilon vitifolium flowering maple M M M / ? ? Healdsburg, Napa, North- San Jose, Salinas, Central 14, 15, 16, 17 1, 2, 3, 4, 6, 8 San Francisco, Coastal San Luis Obispo S Abutilon x hybridum & cvs. flowering maple M H M M / / 1 Auburn, Central Bakersfield, Chico, 8, 9, 14 12, 14, 15, 16 Valley Fresno, Modesto, Sacramento S T Acacia abyssinica Abyssinian acacia / ? / ? / L 2 Irvine, Los South Angeles, Santa 22, 23, 24 1, 2, 4, 6 Coastal Barbara, Ventura, -

Rock Garden Quarterly
ROCK GARDEN QUARTERLY VOLUME 57 NUMBER 4 FALL 1999 This issue of the Rock Garden Quarterly is dedicated to the pre• sent and past Editors of the Chapter newsletters, in gratitude for their commitment to communication and the dissemination of information about rock gardening. In the ten years of my editorship of the Quarterly, amazing strides have been made in the newsletters of the Chapters all across the continent. At first, most newsletters were mere announcements of meetings, and the formats reflected the state of the computers of the time—not too powerful and not too sophisticat• ed. Today, newsletters are full of articles that offer a fund of knowledge, beautifully illustrat• ed, and lovely in format. They spread the word of our art to whole new audiences. Editors, you are training authors not just to contribute to the Quarterly, but to write for many journals and books in the coming century. Thank you to all who have recruited, cajoled, held hands, and written yourselves. Last but not least, thank you to all who have designed, assembled, folded, stamped, and mailed the newsletters, thus making this wonderful resource possible. « 1/ C • C^V COVER: Gentiana sino-ornata by Lori Chips, Norwalk, Connecticut All Material Copyright © 1999 North American Rock Garden Society Printed by AgPress, 1531 Yuma Street, Manhattan, Kansas 66502 ROCK GARDEN QUARTERLY BULLETIN OF THE NORTH AMERICAN ROCK GARDEN SOCIETY VOLUME 57 NUMBER 4 FALL 1999 FEATURES Why Did I Become a Rock Gardener? by Florene Carney 243 Why a Public Rock Garden? by Steve Klass 246 Strategies for Growing Choice Alpines, by Rick Lupp 249 Alliums, by Mark McDonough 261 The Lure of the Orient, by Eva Gallagher 277 Botanizing in Mexico, by Mike Chelednik 285 The Indispensible Blues, by Irma Markert 290 Designing for the Off Season, by Given Kelaidis 293 DEPARTMENTS Awards 297 Book Reviews 302 Winter in the Woodland 307 CARNEY ROCK GARDEN, WASILLA, ALASKA WHY DID I BECOME A ROCK GARDENER? by Florene Carney j^^.