Pollinator Attractors: Petaloidy and Petal Epidermal Cell Shape in Close Relatives of Snapdragon
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Different Speciation Types Meet in a Mediterranean Genus: The
This is an Accepted Manuscript of an article published in Taxon on 4 May 2017, available online: https://doi.org/10.12705/662.7 Different speciation types meet in a Mediterranean genus: the biogeographic history of Cymbalaria (Plantaginaceae). Running head: Phylogeny and biogeographic history of Cymbalaria Pau Carnicero1, Llorenç Sáez1, 2, Núria Garcia-Jacas3 and Mercè Galbany- Casals1 1 Departament de Biologia Animal, Biologia Vegetal i Ecologia, Facultat de Biociències, Universitat Autònoma de Barcelona, 08193 Bellaterra, Spain. 2 Societat d’Història Natural de les Balears (SHNB), C/ Margarida Xirgu 16, E-07011 Palma de Mallorca, Balearic Islands, Spain. 3 Institut Botànic de Barcelona (IBB-CSIC-ICUB), Pg. del Migdia s/n, ES-08038 Barcelona, Spain Author for correspondence: Pau Carnicero, [email protected] ORCID: P.C., http://orcid.org/0000-0002-8345-3309 Abstract Cymbalaria comprises ten species and six subspecies growing in rocky habitats in the Mediterranean Basin. Several features, such as the genus’ highly fragmented distribution as well as noticeable ecological differentiation between partially sympatric species and presence of ploidy barriers between species suggest the involvement of different speciation types in its evolution. The aims of this study were to test the monophyly of Cymbalaria and to reconstruct infrageneric phylogenetic relationships, to infer the genus’ biogeographic history by estimating divergence times and ancestral distribution areas of lineages, and to disentangle the role of different speciation types. To address these issues, we constructed a phylogeny with a complete taxon sampling based on ITS, 3'ETS, ndhF and rpl32-trnL sequences. We used the nuclear ribosomal DNA data to produce a time-calibrated phylogeny, which served as basis for estimating ploidy level evolution and biogeographic history. -

(Balsaminaceae) Using Chloroplast Atpb-Rbcl Spacer Sequences
Systematic Botany (2006), 31(1): pp. 171–180 ᭧ Copyright 2006 by the American Society of Plant Taxonomists Phylogenetics of Impatiens and Hydrocera (Balsaminaceae) Using Chloroplast atpB-rbcL Spacer Sequences STEVEN JANSSENS,1,4 KOEN GEUTEN,1 YONG-MING YUAN,2 YI SONG,2 PHILIPPE KU¨ PFER,2 and ERIK SMETS1,3 1Laboratory of Plant Systematics, Institute of Botany and Microbiology, Kasteelpark Arenberg 31, B-3001 Leuven, Belgium; 2Institut de Botanique, Universite´ de Neuchaˆtel, Neuchaˆtel, Switzerland; 3Nationaal Herbarium Nederland, Universiteit Leiden Branch, PO Box 9514, 2300 RA Leiden, The Netherlands 4Author for correspondence ([email protected]) ABSTRACT. Balsaminaceae are a morphologically diverse family with ca. 1,000 representatives that are mainly distributed in the Old World tropics and subtropics. To understand the relationships of its members, we obtained chloroplast atpB-rbcL sequences from 86 species of Balsaminaceae and five outgroups. Phylogenetic reconstructions using parsimony and Bayesian approaches provide a well-resolved phylogeny in which the sister group relationship between Impatiens and Hydrocera is confirmed. The overall topology of Impatiens is strongly supported and is geographically structured. Impatiens likely origi- nated in South China from which it colonized the adjacent regions and afterwards dispersed into North America, Africa, India, the Southeast Asian peninsula, and the Himalayan region. Balsaminaceae are annual or perennial herbs with in the intrageneric relationships of Impatiens. The most flowers that exhibit a remarkable diversity. The family recent molecular intrageneric study on Balsaminaceae consists of more than 1,000 species (Grey-Wilson utilized Internal Transcribed Spacer (ITS) sequences 1980a; Clifton 2000), but only two genera are recog- and provided new phylogenetic insights in Impatiens nized. -

Vascular Plants and a Brief History of the Kiowa and Rita Blanca National Grasslands
United States Department of Agriculture Vascular Plants and a Brief Forest Service Rocky Mountain History of the Kiowa and Rita Research Station General Technical Report Blanca National Grasslands RMRS-GTR-233 December 2009 Donald L. Hazlett, Michael H. Schiebout, and Paulette L. Ford Hazlett, Donald L.; Schiebout, Michael H.; and Ford, Paulette L. 2009. Vascular plants and a brief history of the Kiowa and Rita Blanca National Grasslands. Gen. Tech. Rep. RMRS- GTR-233. Fort Collins, CO: U.S. Department of Agriculture, Forest Service, Rocky Mountain Research Station. 44 p. Abstract Administered by the USDA Forest Service, the Kiowa and Rita Blanca National Grasslands occupy 230,000 acres of public land extending from northeastern New Mexico into the panhandles of Oklahoma and Texas. A mosaic of topographic features including canyons, plateaus, rolling grasslands and outcrops supports a diverse flora. Eight hundred twenty six (826) species of vascular plant species representing 81 plant families are known to occur on or near these public lands. This report includes a history of the area; ethnobotanical information; an introductory overview of the area including its climate, geology, vegetation, habitats, fauna, and ecological history; and a plant survey and information about the rare, poisonous, and exotic species from the area. A vascular plant checklist of 816 vascular plant taxa in the appendix includes scientific and common names, habitat types, and general distribution data for each species. This list is based on extensive plant collections and available herbarium collections. Authors Donald L. Hazlett is an ethnobotanist, Director of New World Plants and People consulting, and a research associate at the Denver Botanic Gardens, Denver, CO. -

Alien Flora of Europe: Species Diversity, Temporal Trends, Geographical Patterns and Research Needs
Preslia 80: 101–149, 2008 101 Alien flora of Europe: species diversity, temporal trends, geographical patterns and research needs Zavlečená flóra Evropy: druhová diverzita, časové trendy, zákonitosti geografického rozšíření a oblasti budoucího výzkumu Philip W. L a m b d o n1,2#, Petr P y š e k3,4*, Corina B a s n o u5, Martin H e j d a3,4, Margari- taArianoutsou6, Franz E s s l7, Vojtěch J a r o š í k4,3, Jan P e r g l3, Marten W i n t e r8, Paulina A n a s t a s i u9, Pavlos A n d r i opoulos6, Ioannis B a z o s6, Giuseppe Brundu10, Laura C e l e s t i - G r a p o w11, Philippe C h a s s o t12, Pinelopi D e l i p e t - rou13, Melanie J o s e f s s o n14, Salit K a r k15, Stefan K l o t z8, Yannis K o k k o r i s6, Ingolf K ü h n8, Hélia M a r c h a n t e16, Irena P e r g l o v á3, Joan P i n o5, Montserrat Vilà17, Andreas Z i k o s6, David R o y1 & Philip E. H u l m e18 1Centre for Ecology and Hydrology, Hill of Brathens, Banchory, Aberdeenshire AB31 4BW, Scotland, e-mail; [email protected], [email protected]; 2Kew Herbarium, Royal Botanic Gardens Kew, Richmond, Surrey, TW9 3AB, United Kingdom; 3Institute of Bot- any, Academy of Sciences of the Czech Republic, CZ-252 43 Průhonice, Czech Republic, e-mail: [email protected], [email protected], [email protected], [email protected]; 4Department of Ecology, Faculty of Science, Charles University, Viničná 7, CZ-128 01 Praha 2, Czech Republic; e-mail: [email protected]; 5Center for Ecological Research and Forestry Applications, Universitat Autònoma de Barcelona, 08193 Bellaterra, Spain, e-mail: [email protected], [email protected]; 6University of Athens, Faculty of Biology, Department of Ecology & Systematics, 15784 Athens, Greece, e-mail: [email protected], [email protected], [email protected], [email protected], [email protected]; 7Federal Environment Agency, Department of Nature Conservation, Spittelauer Lände 5, 1090 Vienna, Austria, e-mail: [email protected]; 8Helmholtz Centre for Environmental Research – UFZ, Department of Community Ecology, Theodor-Lieser- Str. -

This Thesis Has Been Submitted in Fulfilment of the Requirements for a Postgraduate Degree (E.G
This thesis has been submitted in fulfilment of the requirements for a postgraduate degree (e.g. PhD, MPhil, DClinPsychol) at the University of Edinburgh. Please note the following terms and conditions of use: This work is protected by copyright and other intellectual property rights, which are retained by the thesis author, unless otherwise stated. A copy can be downloaded for personal non-commercial research or study, without prior permission or charge. This thesis cannot be reproduced or quoted extensively from without first obtaining permission in writing from the author. The content must not be changed in any way or sold commercially in any format or medium without the formal permission of the author. When referring to this work, full bibliographic details including the author, title, awarding institution and date of the thesis must be given. Trichome morphology and development in the genus Antirrhinum Ying Tan Doctor of Philosophy Institute of Molecular Plant Sciences School of Biological Sciences The University of Edinburgh 2018 Declaration I declare that this thesis has been composed solely by myself and that it has not been submitted, in whole or in part, in any previous application for a degree. Except where stated otherwise by reference or acknowledgment, the work presented is entirely my own. ___________________ ___________________ Ying Tan Date I Acknowledgments Many people helped and supported me during my study. First, I would like to express my deepest gratitude to my supervisor, Professor Andrew Hudson. He has supported me since my PhD application and always provides his valuable direction and advice. Other members of Prof. Hudson’s research group, especially Erica de Leau and Matthew Barnbrook, taught me lots of experiment skills. -

Turtleheads— the Genus Chelone by Todd Boland, Research Horticulturist, MUNBG
Turtleheads— The Genus Chelone by Todd Boland, Research Horticulturist, MUNBG It seems that most of the flower arrangements. genera of plants grown in our North American gardens hail from Europe or Asia. Only a handful of ornamentals are purely North American natives with Phlox, Rudbeckia and Echinacea as a few examples. Another genus, perhaps not as popular as the species previously mentioned, is Chelone, commonly called turtlehead. In Greek mythology, Chelone was a nymph who made derogatory remarks about the marriage of Zeus and Hera. In retribution, she was turned into a Chelone glabra tortoise, condemning her to eternal silence. Within the Greek language, The most widespread species is the “chelone” then became the word for white turtlehead or balmony, C. tortoise (and hence the common name glabra. This plant extends west as far for this plant). as Manitoba and Minnesota, east to Newfoundland and south as far as There are only 4 species within the Georgia and Mississippi. Rated to zone genus Chelone. In the wild, they grow 3, it is the hardiest species. Although on the edges of streams, lakes, bogs not as showy as the other species, this and wet thickets throughout eastern one does have fragrant flowers. There North America. Based on their growing are at least 9 botanical varieties which preferences in the wild, they prefer a exhibit slightly varying botanical moist site. To accommodate them in differences. The species C. chlorantha the garden, position them in full sun and C. montana, sometimes found in and incorporate plenty of organic older literature, are now classified as material in the growing area as a varieties of the white turtlehead. -
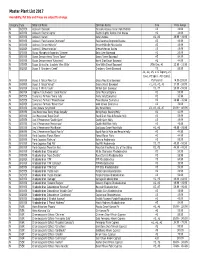
Master Plant List 2017.Xlsx
Master Plant List 2017 Availability, Pot Size and Prices are subject to change. Category Type Botanical Name Common Name Size Price Range N BREVER Azalea X 'Cascade' Cascade Azalea (Glenn Dale Hybrid) #3 49.99 N BREVER Azalea X 'Electric Lights' Electric Lights Double Pink Azalea #2 44.99 N BREVER Azalea X 'Karen' Karen Azalea #2, #3 39.99 - 49.99 N BREVER Azalea X 'Poukhanense Improved' Poukhanense Improved Azalea #3 49.99 N BREVER Azalea X 'Renee Michelle' Renee Michelle Pink Azalea #3 49.99 N BREVER Azalea X 'Stewartstonian' Stewartstonian Azalea #3 49.99 N BREVER Buxus Microphylla Japonica "Gregem' Baby Gem Boxwood #2 29.99 N BREVER Buxus Sempervirens 'Green Tower' Green Tower Boxwood #5 64.99 N BREVER Buxus Sempervirens 'Katerberg' North Star Dwarf Boxwood #2 44.99 N BREVER Buxus Sinica Var. Insularis 'Wee Willie' Wee Willie Dwarf Boxwood Little One, #1 13.99 - 21.99 N BREVER Buxus X 'Cranberry Creek' Cranberry Creek Boxwood #3 89.99 #1, #2, #5, #15 Topiary, #5 Cone, #5 Spiral, #10 Spiral, N BREVER Buxus X 'Green Mountain' Green Mountain Boxwood #5 Pyramid 14.99-299.99 N BREVER Buxus X 'Green Velvet' Green Velvet Boxwood #1, #2, #3, #5 17.99 - 59.99 N BREVER Buxus X 'Winter Gem' Winter Gem Boxwood #5, #7 59.99 - 99.99 N BREVER Daphne X Burkwoodii 'Carol Mackie' Carol Mackie Daphne #2 59.99 N BREVER Euonymus Fortunei 'Ivory Jade' Ivory Jade Euonymus #2 35.99 N BREVER Euonymus Fortunei 'Moonshadow' Moonshadow Euonymus #2 29.99 - 35.99 N BREVER Euonymus Fortunei 'Rosemrtwo' Gold Splash Euonymus #2 39.99 N BREVER Ilex Crenata 'Sky Pencil' -

Flora Mediterranea 26
FLORA MEDITERRANEA 26 Published under the auspices of OPTIMA by the Herbarium Mediterraneum Panormitanum Palermo – 2016 FLORA MEDITERRANEA Edited on behalf of the International Foundation pro Herbario Mediterraneo by Francesco M. Raimondo, Werner Greuter & Gianniantonio Domina Editorial board G. Domina (Palermo), F. Garbari (Pisa), W. Greuter (Berlin), S. L. Jury (Reading), G. Kamari (Patras), P. Mazzola (Palermo), S. Pignatti (Roma), F. M. Raimondo (Palermo), C. Salmeri (Palermo), B. Valdés (Sevilla), G. Venturella (Palermo). Advisory Committee P. V. Arrigoni (Firenze) P. Küpfer (Neuchatel) H. M. Burdet (Genève) J. Mathez (Montpellier) A. Carapezza (Palermo) G. Moggi (Firenze) C. D. K. Cook (Zurich) E. Nardi (Firenze) R. Courtecuisse (Lille) P. L. Nimis (Trieste) V. Demoulin (Liège) D. Phitos (Patras) F. Ehrendorfer (Wien) L. Poldini (Trieste) M. Erben (Munchen) R. M. Ros Espín (Murcia) G. Giaccone (Catania) A. Strid (Copenhagen) V. H. Heywood (Reading) B. Zimmer (Berlin) Editorial Office Editorial assistance: A. M. Mannino Editorial secretariat: V. Spadaro & P. Campisi Layout & Tecnical editing: E. Di Gristina & F. La Sorte Design: V. Magro & L. C. Raimondo Redazione di "Flora Mediterranea" Herbarium Mediterraneum Panormitanum, Università di Palermo Via Lincoln, 2 I-90133 Palermo, Italy [email protected] Printed by Luxograph s.r.l., Piazza Bartolomeo da Messina, 2/E - Palermo Registration at Tribunale di Palermo, no. 27 of 12 July 1991 ISSN: 1120-4052 printed, 2240-4538 online DOI: 10.7320/FlMedit26.001 Copyright © by International Foundation pro Herbario Mediterraneo, Palermo Contents V. Hugonnot & L. Chavoutier: A modern record of one of the rarest European mosses, Ptychomitrium incurvum (Ptychomitriaceae), in Eastern Pyrenees, France . 5 P. Chène, M. -

Baja California, Mexico, and a Vegetation Map of Colonet Mesa Alan B
Aliso: A Journal of Systematic and Evolutionary Botany Volume 29 | Issue 1 Article 4 2011 Plants of the Colonet Region, Baja California, Mexico, and a Vegetation Map of Colonet Mesa Alan B. Harper Terra Peninsular, Coronado, California Sula Vanderplank Rancho Santa Ana Botanic Garden, Claremont, California Mark Dodero Recon Environmental Inc., San Diego, California Sergio Mata Terra Peninsular, Coronado, California Jorge Ochoa Long Beach City College, Long Beach, California Follow this and additional works at: http://scholarship.claremont.edu/aliso Part of the Biodiversity Commons, Botany Commons, and the Ecology and Evolutionary Biology Commons Recommended Citation Harper, Alan B.; Vanderplank, Sula; Dodero, Mark; Mata, Sergio; and Ochoa, Jorge (2011) "Plants of the Colonet Region, Baja California, Mexico, and a Vegetation Map of Colonet Mesa," Aliso: A Journal of Systematic and Evolutionary Botany: Vol. 29: Iss. 1, Article 4. Available at: http://scholarship.claremont.edu/aliso/vol29/iss1/4 Aliso, 29(1), pp. 25–42 ’ 2011, Rancho Santa Ana Botanic Garden PLANTS OF THE COLONET REGION, BAJA CALIFORNIA, MEXICO, AND A VEGETATION MAPOF COLONET MESA ALAN B. HARPER,1 SULA VANDERPLANK,2 MARK DODERO,3 SERGIO MATA,1 AND JORGE OCHOA4 1Terra Peninsular, A.C., PMB 189003, Suite 88, Coronado, California 92178, USA ([email protected]); 2Rancho Santa Ana Botanic Garden, 1500 North College Avenue, Claremont, California 91711, USA; 3Recon Environmental Inc., 1927 Fifth Avenue, San Diego, California 92101, USA; 4Long Beach City College, 1305 East Pacific Coast Highway, Long Beach, California 90806, USA ABSTRACT The Colonet region is located at the southern end of the California Floristic Province, in an area known to have the highest plant diversity in Baja California. -

Plants, Volume 1, Number 1 (August 1979)
Desert Plants, Volume 1, Number 1 (August 1979) Item Type Article Publisher University of Arizona (Tucson, AZ) Journal Desert Plants Rights Copyright © Arizona Board of Regents. The University of Arizona. Download date 02/10/2021 01:18:53 Link to Item http://hdl.handle.net/10150/528188 Volume I. Number 1. August 1979 Desert Published by The University of Arizona for the Plants Boyce Thompson Southwestern Arboretum Assisting Nature with Plant Selection4 Larry K. Holzworth Aberrant Sex -Ratios in Jojoba Associated with Environmental Factors 8 Serena L. Cole 'J. G. Lemmon & Wife,' Plant Explorers in Arizona, California, and Nevada12 Frank S. Crosswhite 'Extinct' Wire -Lettuce, Stephanomeria schottii (Compositae), Rediscovered in Arizona after More Than One Hundred Years22 Elinor Lehto Southwestern Indian Sunflowers23 Gary Paul Nabhan Transition from a Bermudagrass Lawn to a Landscape of Rock or Gravel Mulch 27 Charles Sacamano Preliminary Evaluation of Cold- hardiness in Desert Landscaping Plants at Central Arizona College29 William A. Kinnison Effects of the 1978 Freeze on Native Plants of Sonora, Mexico33 Warren D. Jones The Severe Freeze of 1978 -79 in the Southwestern United States37 The National Climate Program Act of 197840 Reviews42 Arboretum Progress46 R. T. McKittrick Volume 1. Number 1. August 1979 Published by The University of Arizona Desert Plants for the Boyce Thompson Southwestern Arboretum The Severe Freeze of 1978 -79 in the Contents Southwestern United States37 Correspondents: Editorial Barrie D. Coate, Saratoga Horticultural Foundation; Dara E. Emery, Santa Barbara Botanic Garden; Louis C. Assisting Nature with Plant Selection 4 Erickson, Botanic Gardens, University of California, River- Larry K. Holzworth, USDA Soil Conservation side; Wayne L. -

Diversity and Evolution of Asterids!
Diversity and Evolution of Asterids! . mints and snapdragons . ! *Boraginaceae - borage family! Widely distributed, large family of alternate leaved plants. Typically hairy. Typically possess helicoid or scorpiod cymes = compound monochasium. Many are poisonous or used medicinally. Mertensia virginica - Eastern bluebells *Boraginaceae - borage family! CA (5) CO (5) A 5 G (2) Gynobasic style; not terminal style which is usual in plants; this feature is shared with the mint family (Lamiaceae) which is not related Myosotis - forget me not 2 carpels each with 2 ovules are separated at maturity and each further separated into 1 ovuled compartments Fruit typically 4 nutlets *Boraginaceae - borage family! Echium vulgare Blueweed, viper’s bugloss adventive *Boraginaceae - borage family! Hackelia virginiana Beggar’s-lice Myosotis scorpioides Common forget-me-not *Boraginaceae - borage family! Lithospermum canescens Lithospermum incisium Hoary puccoon Fringed puccoon *Boraginaceae - borage family! pin thrum Lithospermum canescens • Lithospermum (puccoon) - classic Hoary puccoon dimorphic heterostyly *Boraginaceae - borage family! Mertensia virginica Eastern bluebells Botany 401 final field exam plant! *Boraginaceae - borage family! Leaves compound or lobed and “water-marked” Hydrophyllum virginianum - Common waterleaf Botany 401 final field exam plant! **Oleaceae - olive family! CA (4) CO (4) or 0 A 2 G (2) • Woody plants, opposite leaves • 4 merous actinomorphic or regular flowers Syringa vulgaris - Lilac cultivated **Oleaceae - olive family! CA (4) -

1 Executive Summary the Annual
Taylor’s Checkerspot Butterfly Working Group Annual Meeting | Nisqually Wildlife Refuge | Nov. 6th & 7th, 2013 Executive Summary The annual Taylor’s checkerspot butterfly (TCB) working group meeting includes updates to research, monitoring, and conservation actions as well as a needs discussion and update to the Action Plan that lists and prioritizes next best actions for Taylor’s that can be accomplished in the next 3-5 years. One major update from the 2013 meeting was the federal listing of the Taylor’s checkerspot butterfly as endangered under the ESA. In addition to this major change in the regulatory landscape, many partners have continued to make progress in land protection and restoration, monitoring, and reintroduction efforts, with much of the work being done collaboratively among multiple partners. This year’s meeting also had multiple presentations of preliminary research results, which allowed for interesting discussion about how to translate knowledge learned through lab and field research to on-the-ground management changes and decisions. Day 1 of the meeting included specific project/initiative updates. Ted Thomas (USFWS) provided an overview of impacts and opportunities resulting from the recent ESA listing of the Taylor’s checkerspot butterfly as endangered, including critical habitat designation. Following this, two researchers presented data about habitat quality, host plants, and the role this information can play in recovery efforts. Elizabeth Crone (Tufts) presented her research on identifying if ‘bad’ habitat can be beneficial – finding it can play a positive role - and the different ways in which butterflies use suitable and less-suitable habitat. Nate Haan (UW) presented preliminary results from his research on the history of the relationship between golden paintbrush and TCB, host-plant fidelity, and how this research can improve our reintroduction efforts.