A002 Methylobacterium Carri Sp. Nov., Isolated from Automotive Air
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The 2014 Golden Gate National Parks Bioblitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event
National Park Service U.S. Department of the Interior Natural Resource Stewardship and Science The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 ON THIS PAGE Photograph of BioBlitz participants conducting data entry into iNaturalist. Photograph courtesy of the National Park Service. ON THE COVER Photograph of BioBlitz participants collecting aquatic species data in the Presidio of San Francisco. Photograph courtesy of National Park Service. The 2014 Golden Gate National Parks BioBlitz - Data Management and the Event Species List Achieving a Quality Dataset from a Large Scale Event Natural Resource Report NPS/GOGA/NRR—2016/1147 Elizabeth Edson1, Michelle O’Herron1, Alison Forrestel2, Daniel George3 1Golden Gate Parks Conservancy Building 201 Fort Mason San Francisco, CA 94129 2National Park Service. Golden Gate National Recreation Area Fort Cronkhite, Bldg. 1061 Sausalito, CA 94965 3National Park Service. San Francisco Bay Area Network Inventory & Monitoring Program Manager Fort Cronkhite, Bldg. 1063 Sausalito, CA 94965 March 2016 U.S. Department of the Interior National Park Service Natural Resource Stewardship and Science Fort Collins, Colorado The National Park Service, Natural Resource Stewardship and Science office in Fort Collins, Colorado, publishes a range of reports that address natural resource topics. These reports are of interest and applicability to a broad audience in the National Park Service and others in natural resource management, including scientists, conservation and environmental constituencies, and the public. The Natural Resource Report Series is used to disseminate comprehensive information and analysis about natural resources and related topics concerning lands managed by the National Park Service. -

Ecology and Epidemiology of Campylobacter Jejuni in Broiler Chickens
Ecology and Epidemiology of Campylobacter jejuni in Broiler Chickens A DISSERTATION SUBMITTED TO THE FACULTY OF THE UNIVERSITY OF MINNESOTA BY Hae Jin Hwang IN PARTIALL FULFILLMENT OF THE REQUIREMENTS FOR THE DEGREE OF DOCTOR OF PHILOSOPHY Dr. Randall Singer, Dr. George Maldonado June 2019 © Hae Jin Hwang, 2019 Acknowledgements I would like to sincerely thank my advisor, Dr. Randall Singer, for his intellectual guidance and support, great patience, and mentorship, which made this dissertation possible. I would also like to thank Dr. George Maldonado for his continuous encouragement and support. I would further like to thank my thesis committee, Dr. Richard Isaacson and Dr. Timothy Church, for their guidance throughout my doctoral training. I thank all my friends and colleagues I met over the course of my studies. I am especially indebted to my friends, Dr. Kristy Lee, Dr. Irene Bueno Padilla, Dr. Elise Lamont, Madhumathi Thiruvengadam, Dr. Kaushi Kanankege and Dr. Sylvia Wanzala, for their support and friendship. Heartfelt gratitude goes to my family, for always believing in me, encouraging me and helping me get through the difficult and stressful times during my studies. Lastly, I thank Sven and Bami for being the best writing companions I could ever ask for. i Abstract Campylobacteriosis, predominantly caused by Campylobacter jejuni, is a common, yet serious foodborne illness. With consumption and handling of poultry products as the most important risk factor of campylobacteriosis, reducing Campylobacter contamination in poultry products is considered the best public health intervention to reduce the burden and costs associated with campylobacteriosis. To this end, there is a need to improve our understanding of epidemiology and ecology of Campylobacter jejuni in poultry. -

Elisabeth Mendes Martins De Moura Diversidade De Vírus DNA
Elisabeth Mendes Martins de Moura Diversidade de vírus DNA autóctones e alóctones de mananciais e de esgoto da região metropolitana de São Paulo Tese apresentada ao Programa de Pós- Graduação em Microbiologia do Instituto de Ciências Biomédicas da Universidade de São Paulo, para obtenção do Titulo de Doutor em Ciências. Área de concentração: Microbiologia Orienta: Prof (a). Dr (a). Dolores Ursula Mehnert versão original São Paulo 2017 RESUMO MOURA, E. M. M. Diversidade de vírus DNA autóctones e alóctones de mananciais e de esgoto da região metropolitana de São Paulo. 2017. 134f. Tese (Doutorado em Microbiologia) - Instituto de Ciências Biomédicas, Universidade de São Paulo, São Paulo, 2017. A água doce no Brasil, assim como o seu consumo é extremamente importante para as diversas atividades criadas pelo ser humano. Por esta razão o consumo deste bem é muito grande e consequentemente, provocando o seu impacto. Os mananciais são normalmente usados para abastecimento doméstico, comercial, industrial e outros fins. Os estudos na área de ecologia de micro-organismos nos ecossistemas aquáticos (mananciais) e em esgotos vêm sendo realizados com mais intensidade nos últimos anos. Nas últimas décadas foi introduzido o conceito de virioplâncton com base na abundância e diversidade de partículas virais presentes no ambiente aquático. O virioplâncton influencia muitos processos ecológicos e biogeoquímicos, como ciclagem de nutriente, taxa de sedimentação de partículas, diversidade e distribuição de espécies de algas e bactérias, controle de florações de fitoplâncton e transferência genética horizontal. Os estudos nesta área da virologia molecular ainda estão muito restritos no país, bem como muito pouco se conhece sobre a diversidade viral na água no Brasil. -

A Taxonomic Note on the Genus Lactobacillus
Taxonomic Description template 1 A taxonomic note on the genus Lactobacillus: 2 Description of 23 novel genera, emended description 3 of the genus Lactobacillus Beijerinck 1901, and union 4 of Lactobacillaceae and Leuconostocaceae 5 Jinshui Zheng1, $, Stijn Wittouck2, $, Elisa Salvetti3, $, Charles M.A.P. Franz4, Hugh M.B. Harris5, Paola 6 Mattarelli6, Paul W. O’Toole5, Bruno Pot7, Peter Vandamme8, Jens Walter9, 10, Koichi Watanabe11, 12, 7 Sander Wuyts2, Giovanna E. Felis3, #*, Michael G. Gänzle9, 13#*, Sarah Lebeer2 # 8 '© [Jinshui Zheng, Stijn Wittouck, Elisa Salvetti, Charles M.A.P. Franz, Hugh M.B. Harris, Paola 9 Mattarelli, Paul W. O’Toole, Bruno Pot, Peter Vandamme, Jens Walter, Koichi Watanabe, Sander 10 Wuyts, Giovanna E. Felis, Michael G. Gänzle, Sarah Lebeer]. 11 The definitive peer reviewed, edited version of this article is published in International Journal of 12 Systematic and Evolutionary Microbiology, https://doi.org/10.1099/ijsem.0.004107 13 1Huazhong Agricultural University, State Key Laboratory of Agricultural Microbiology, Hubei Key 14 Laboratory of Agricultural Bioinformatics, Wuhan, Hubei, P.R. China. 15 2Research Group Environmental Ecology and Applied Microbiology, Department of Bioscience 16 Engineering, University of Antwerp, Antwerp, Belgium 17 3 Dept. of Biotechnology, University of Verona, Verona, Italy 18 4 Max Rubner‐Institut, Department of Microbiology and Biotechnology, Kiel, Germany 19 5 School of Microbiology & APC Microbiome Ireland, University College Cork, Co. Cork, Ireland 20 6 University of Bologna, Dept. of Agricultural and Food Sciences, Bologna, Italy 21 7 Research Group of Industrial Microbiology and Food Biotechnology (IMDO), Vrije Universiteit 22 Brussel, Brussels, Belgium 23 8 Laboratory of Microbiology, Department of Biochemistry and Microbiology, Ghent University, Ghent, 24 Belgium 25 9 Department of Agricultural, Food & Nutritional Science, University of Alberta, Edmonton, Canada 26 10 Department of Biological Sciences, University of Alberta, Edmonton, Canada 27 11 National Taiwan University, Dept. -

Scope of the Thesis
Unraveling predatory-prey interactions between bacteria Dissertation To Fulfill the Requirements for the Degree of „Doctor rerum naturalium“ (Dr. rer. nat.) Submitted to the Council of the Faculty of Biology and Pharmacy of the Friedrich Schiller University Jena By Ivana Seccareccia (M.Sc. in Biology) born on 8th April 1986 in Rijeka, Croatia Die Forschungsarbeit im Rahmen dieser Dissertation wurde am Leibniz-Institut für Naturstoff-Forschung und Infektionsbiologie e.V. – Hans-Knöll–Institut in der Nachwuchsgruppe Sekundärmetabolismus räuberischer Bakterien unter der Betreuung von Dr. habil. Markus Nett von Oktober 2011 bis Oktober 2015 in Jena durchgeführt. Gutachter: ……………………………………………. ………………………………………….… ……………………………………………. Tag der öffentlichen Verteidigung: We make our world significant by the courage of our questions and by the depth of our answers. Carl Sagan Table of Contents 1 Introduction ......................................................................................................................... 6 1.1 Predation in the microbial community ........................................................................... 6 1.2 Bacterial predators .......................................................................................................... 7 1.3 Phases of predation ......................................................................................................... 8 1.3.1 Seeking prey ......................................................................................................... 9 1.3.2 Prey recognition -

Lysobacter Enzymogenes
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Dissertations and Theses in Biological Sciences Biological Sciences, School of 8-2010 Detection Methods for the Genus Lysobacter and the Species Lysobacter enzymogenes Hu Yin University of Nebraska at Lincoln, [email protected] Follow this and additional works at: https://digitalcommons.unl.edu/bioscidiss Part of the Life Sciences Commons Yin, Hu, "Detection Methods for the Genus Lysobacter and the Species Lysobacter enzymogenes" (2010). Dissertations and Theses in Biological Sciences. 15. https://digitalcommons.unl.edu/bioscidiss/15 This Article is brought to you for free and open access by the Biological Sciences, School of at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Dissertations and Theses in Biological Sciences by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Detection Methods for the Genus Lysobacter and the Species Lysobacter enzymogenes By Hu Yin A THESIS Presented to the Faculty of The Graduate College at the University of Nebraska In Partial Fulfillment of Requirements For the Degree of Master of Science Major: Biological Sciences Under the Supervision of Professor Gary Y. Yuen Lincoln, Nebraska August, 2010 Detection Methods for the Genus Lysobacter and the Species Lysobacter enzymogenes Hu Yin, M.S. University of Nebraska, 2010 Advisor: Gary Y. Yuen Strains of Lysobacter enzymogenes, a bacterial species with biocontrol activity, have been detected via 16S rDNA sequences in soil in different parts of the world. In most instances, however, their occurrence could not be confirmed by isolation, presumably because the species occurred in low numbers relative to faster-growing species of Bacillus or Pseudomonas. -
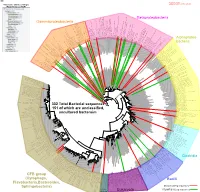
Bacteria Clostridia Bacilli Eukaryota CFB Group
AM935842.1.1361 uncultured Burkholderiales bacterium Class Betaproteobacteria AY283260.1.1552 Alcaligenes sp. PCNB−2 Class Betaproteobacteria AM934953.1.1374 uncultured Burkholderiales bacterium Class Betaproteobacteria AJ581593.1.1460 uncultured betaAM936569.1.1351 proteobacterium uncultured Class Betaproteobacteria Derxia sp. Class Betaproteobacteria AJ581621.1.1418 uncultured beta proteobacterium Class Betaproteobacteria DQ248272.1.1498 uncultured soil bacterium soil uncultured DQ248272.1.1498 DQ248235.1.1498 uncultured soil bacterium RS49 DQ248270.1.1496 uncultured soil bacterium DQ256489.1.1211 Variovorax paradoxus Class Betaproteobacteria Class paradoxus Variovorax DQ256489.1.1211 AF523053.1.1486 uncultured Comamonadaceae bacterium Class Betaproteobacteria AY706442.1.1396 uncultured bacterium uncultured AY706442.1.1396 AJ536763.1.1422 uncultured bacterium CS000359.1.1530 Variovorax paradoxus Class Betaproteobacteria Class paradoxus Variovorax CS000359.1.1530 AY168733.1.1411 uncultured bacterium AJ009470.1.1526 uncultured bacterium SJA−62 Class Betaproteobacteria Class SJA−62 bacterium uncultured AJ009470.1.1526 AY212561.1.1433 uncultured bacterium D16212.1.1457 Rhodoferax fermentans Class Betaproteobacteria Class fermentans Rhodoferax D16212.1.1457 AY957894.1.1546 uncultured bacterium AJ581620.1.1452 uncultured beta proteobacterium Class Betaproteobacteria RS76 AY625146.1.1498 uncultured bacterium RS65 DQ316832.1.1269 uncultured beta proteobacterium Class Betaproteobacteria DQ404909.1.1513 uncultured bacterium uncultured DQ404909.1.1513 AB021341.1.1466 bacterium rM6 AJ487020.1.1500 uncultured bacterium uncultured AJ487020.1.1500 RS7 RS86RC AF364862.1.1425 bacterium BA128 Class Betaproteobacteria AY957931.1.1529 uncultured bacterium uncultured AY957931.1.1529 CP000884.723807.725332 Delftia acidovorans SPH−1 Class Betaproteobacteria AY957923.1.1520 uncultured bacterium uncultured AY957923.1.1520 RS18 AY957918.1.1527 uncultured bacterium uncultured AY957918.1.1527 AY945883.1.1500 uncultured bacterium AF526940.1.1489 uncultured Ralstonia sp. -

Virus World As an Evolutionary Network of Viruses and Capsidless Selfish Elements
Virus World as an Evolutionary Network of Viruses and Capsidless Selfish Elements Koonin, E. V., & Dolja, V. V. (2014). Virus World as an Evolutionary Network of Viruses and Capsidless Selfish Elements. Microbiology and Molecular Biology Reviews, 78(2), 278-303. doi:10.1128/MMBR.00049-13 10.1128/MMBR.00049-13 American Society for Microbiology Version of Record http://cdss.library.oregonstate.edu/sa-termsofuse Virus World as an Evolutionary Network of Viruses and Capsidless Selfish Elements Eugene V. Koonin,a Valerian V. Doljab National Center for Biotechnology Information, National Library of Medicine, Bethesda, Maryland, USAa; Department of Botany and Plant Pathology and Center for Genome Research and Biocomputing, Oregon State University, Corvallis, Oregon, USAb Downloaded from SUMMARY ..................................................................................................................................................278 INTRODUCTION ............................................................................................................................................278 PREVALENCE OF REPLICATION SYSTEM COMPONENTS COMPARED TO CAPSID PROTEINS AMONG VIRUS HALLMARK GENES.......................279 CLASSIFICATION OF VIRUSES BY REPLICATION-EXPRESSION STRATEGY: TYPICAL VIRUSES AND CAPSIDLESS FORMS ................................279 EVOLUTIONARY RELATIONSHIPS BETWEEN VIRUSES AND CAPSIDLESS VIRUS-LIKE GENETIC ELEMENTS ..............................................280 Capsidless Derivatives of Positive-Strand RNA Viruses....................................................................................................280 -

Exploring the Tymovirids Landscape Through Metatranscriptomics Data
bioRxiv preprint doi: https://doi.org/10.1101/2021.07.15.452586; this version posted July 16, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Exploring the tymovirids landscape through metatranscriptomics data 2 Nicolás Bejerman1,2, Humberto Debat1,2 3 4 1 Instituto de Patología Vegetal – Centro de Investigaciones Agropecuarias – Instituto Nacional de 5 Tecnología Agropecuaria (IPAVE-CIAP-INTA), Camino 60 Cuadras Km 5,5 (X5020ICA), Córdoba, 6 Argentina 7 2 Consejo Nacional de Investigaciones Científicas y Técnicas. Unidad de Fitopatología y Modelización 8 Agrícola, Camino 60 Cuadras Km 5,5 (X5020ICA), Córdoba, Argentina 9 10 Corresponding author: Nicolás Bejerman, [email protected] 11 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.07.15.452586; this version posted July 16, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 12 Abstract 13 Tymovirales is an order of viruses with positive-sense, single-stranded RNA genomes that mostly infect 14 plants, but also fungi and insects. The number of tymovirid sequences has been growing in the last few 15 years with the extensive use of high-throughput sequencing platforms. Here we report the discovery of 31 16 novel tymovirid genomes associated with 27 different host plant species, which were hidden in public 17 databases. -

Melbourne Australia 16 – 19 November 2010
9th h 9t MelbourneMelbourne AustraliaAustralia 1616 –– 1919 NovemberNovember 20102010 Sponsored by ISBN 978-1-74264-590-2 (online) Welcome Welcome to the 9th Australasian Plant Virology Workshop (APVW). The organising committee would like to thank our fellow plant virologists and researchers of virus-like organisms for attending our virology workshop in Melbourne. This workshop provides a forum for plant virologists from Australia, New Zealand and the rest of the world to meet on a biannual basis and discuss plant virus and virus-like topics in a congenial environment, over four days. Our theme for the workshop is “Plant viruses: Friend or foe?” We would like to build on the concepts introduced to this group at the 8th Australasian Plant Virology Workshop and consider the role of viruses as both pathogens and as beneficial organisms. The talks and posters for this workshop have been aligned with seven themes: Plant virus ecology and diversity; New and emerging viruses; Virus-like organisms; Plant virus diversity and detection; New tools and technologies; Plant host-virus interactions; and Plant virus epidemiology and climate change. We have officially aligned the APVW with the Australasian Plant Pathology Society as a special interest group and we now have an official website aligned with APPS (http://www.australasianplantpathologysociety.org.au/). As part of the program we have also scheduled a one hour meeting to discuss the formalisation of the Australasian Plant Virology Special Interest Group (APV) and any issues that are relevant to our newly formed group. We would like to extend a special thanks to several organisations for their support: • Horticulture Australia Limited (HAL) for sponsoring this workshop • Cooperative Research Centre for National Plant Biosecurity who are a platinum sponsor of this workshop, and providing the satchels • Victorian Department of Primary Industries who sponsored Ulrich Melcher and Ko Verhoeven • Qiagen as an exhibition sponsor, and • The Jasper Hotel for their support in hosting the workshop. -

Contamination of Sulfonamide Antibiotics and Sulfamethazine-Resistant Bacteria in the Downstream and Estuarine Areas of Jiulong River in Southeast China
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Crossref Environ Sci Pollut Res (2015) 22:12104–12113 DOI 10.1007/s11356-015-4473-z RESEARCH ARTICLE Contamination of sulfonamide antibiotics and sulfamethazine-resistant bacteria in the downstream and estuarine areas of Jiulong River in Southeast China Danyun Ou1 & Bin Chen1 & Renao Bai 1 & Puqing Song1 & Heshan Lin1 Received: 28 December 2014 /Accepted: 30 March 2015 /Published online: 16 April 2015 # The Author(s) 2015. This article is published with open access at Springerlink.com Abstract Surface water samples from downstream and estu- Introduction arine areas of Jiulong River were collected in August 2011 and May 2012 for detecting sulfonamide antibiotic residues The occurrence of antibiotics in aquatic environments is of and isolating sulfamethazine-resistant bacteria. ecotoxicological concern because of potential ecosystem al- Sulfamethazine was detected in all samples in May 2012 at teration. Prolonged exposure to low doses of antibiotics leads an average concentration of 78.3 ng L−1, which was the to selective proliferation of resistant bacteria, which could highest among the nine sulfonamide antibiotics determined. transfer antimicrobial resistance genes to other bacterial spe- Sulfamethazine-resistant bacteria (SRB) were screened using cies (Dantas et al. 2008;LevyandMarshall2004)with antibiotic-containing agar plates. The SRB average abundance difficult-to-predict consequences on human health (Martínez in the samples was 3.69×104 and 2.17×103 CFUs mL−1 in 2008). The potential of these low antibiotic concentrations to August 2011 and May 2012, respectively, and was positively promote antimicrobial resistance has been substantiated by the correlated to sulfamethazine concentration in May 2012. -

Insights Into 6S RNA in Lactic Acid Bacteria (LAB) Pablo Gabriel Cataldo1,Paulklemm2, Marietta Thüring2, Lucila Saavedra1, Elvira Maria Hebert1, Roland K
Cataldo et al. BMC Genomic Data (2021) 22:29 BMC Genomic Data https://doi.org/10.1186/s12863-021-00983-2 RESEARCH ARTICLE Open Access Insights into 6S RNA in lactic acid bacteria (LAB) Pablo Gabriel Cataldo1,PaulKlemm2, Marietta Thüring2, Lucila Saavedra1, Elvira Maria Hebert1, Roland K. Hartmann2 and Marcus Lechner2,3* Abstract Background: 6S RNA is a regulator of cellular transcription that tunes the metabolism of cells. This small non-coding RNA is found in nearly all bacteria and among the most abundant transcripts. Lactic acid bacteria (LAB) constitute a group of microorganisms with strong biotechnological relevance, often exploited as starter cultures for industrial products through fermentation. Some strains are used as probiotics while others represent potential pathogens. Occasional reports of 6S RNA within this group already indicate striking metabolic implications. A conceivable idea is that LAB with 6S RNA defects may metabolize nutrients faster, as inferred from studies of Echerichia coli.Thismay accelerate fermentation processes with the potential to reduce production costs. Similarly, elevated levels of secondary metabolites might be produced. Evidence for this possibility comes from preliminary findings regarding the production of surfactin in Bacillus subtilis, which has functions similar to those of bacteriocins. The prerequisite for its potential biotechnological utility is a general characterization of 6S RNA in LAB. Results: We provide a genomic annotation of 6S RNA throughout the Lactobacillales order. It laid the foundation for a bioinformatic characterization of common 6S RNA features. This covers secondary structures, synteny, phylogeny, and product RNA start sites. The canonical 6S RNA structure is formed by a central bulge flanked by helical arms and a template site for product RNA synthesis.