Mitigating the Inhibition of Human Bile Salt Export Pump by Drugs
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
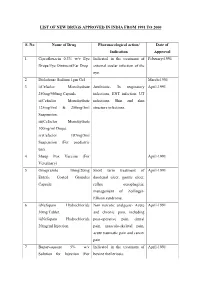
List of New Drugs Approved in India from 1991 to 2000
LIST OF NEW DRUGS APPROVED IN INDIA FROM 1991 TO 2000 S. No Name of Drug Pharmacological action/ Date of Indication Approval 1 Ciprofloxacin 0.3% w/v Eye Indicated in the treatment of February-1991 Drops/Eye Ointment/Ear Drop external ocular infection of the eye. 2 Diclofenac Sodium 1gm Gel March-1991 3 i)Cefaclor Monohydrate Antibiotic- In respiratory April-1991 250mg/500mg Capsule. infections, ENT infection, UT ii)Cefaclor Monohydrate infections, Skin and skin 125mg/5ml & 250mg/5ml structure infections. Suspension. iii)Cefaclor Monohydrate 100mg/ml Drops. iv)Cefaclor 187mg/5ml Suspension (For paediatric use). 4 Sheep Pox Vaccine (For April-1991 Veterinary) 5 Omeprazole 10mg/20mg Short term treatment of April-1991 Enteric Coated Granules duodenal ulcer, gastric ulcer, Capsule reflux oesophagitis, management of Zollinger- Ellison syndrome. 6 i)Nefopam Hydrochloride Non narcotic analgesic- Acute April-1991 30mg Tablet. and chronic pain, including ii)Nefopam Hydrochloride post-operative pain, dental 20mg/ml Injection. pain, musculo-skeletal pain, acute traumatic pain and cancer pain. 7 Buparvaquone 5% w/v Indicated in the treatment of April-1991 Solution for Injection (For bovine theileriosis. Veterinary) 8 i)Kitotifen Fumerate 1mg Anti asthmatic drug- Indicated May-1991 Tablet in prophylactic treatment of ii)Kitotifen Fumerate Syrup bronchial asthma, symptomatic iii)Ketotifen Fumerate Nasal improvement of allergic Drops conditions including rhinitis and conjunctivitis. 9 i)Pefloxacin Mesylate Antibacterial- In the treatment May-1991 Dihydrate 400mg Film Coated of severe infection in adults Tablet caused by sensitive ii)Pefloxacin Mesylate microorganism (gram -ve Dihydrate 400mg/5ml Injection pathogens and staphylococci). iii)Pefloxacin Mesylate Dihydrate 400mg I.V Bottles of 100ml/200ml 10 Ofloxacin 100mg/50ml & Indicated in RTI, UTI, May-1991 200mg/100ml vial Infusion gynaecological infection, skin/soft lesion infection. -
ALPHA ADRENOCEPTORS and HUMAN SEXUAL FUNCTION Alan
8 1995 Elsevier Science B. V. All rights reserved. The Pharmacology of Sexual Function and Dysfunction J. Bancroft, editor 307 ALPHA ADRENOCEPTORS AND HUMAN SEXUAL FUNCTION Alan J Riley Field Place, Dunsmore, Buckinghamshire, HP22 6QH, UK Introduction Sexual functioning involves complex physiological processes which rely on the interplay of many central and peripheral neurotransmitter systems. Disturbances in any one of these systems might be associated with disturbed sexual function which, when recognised, may be alleviated by appropriate pharmacological manipulation, although at the present time this is more hypothesis than reality, The sympathetic nervous system is involved actively at various levels in the normal control of sexual responses. The effects of sympathetic activation are mediated by the release of noradrenaline from nerve terminals and the increased secretion of adrenaline from the adrenal medulla. These catecholamines selectively activate specific cellular sites in target tissues known as adrenoceptors (previously termed adrenergic receptors) to mediate responses. Almost fifty years ago, Alquist realised that tissue responses to catecholamines were mediated through two distinct types of receptors which he designated a and/? [1]. This review focuses on the involvement of a-adrenoceptors in human sexual functioning and dysfunction. Alpha adrenoceptors are located both pre- and post- synaptically and they were classified as either ar or af adrenoceptors according to location; or, being postsynaptic and az presynaptic. This classification continues to be used in some texts. However, as highly specific and selective pharmacological tools became available, this locational subclassification is found not always to be appropriate. Nowadays, classification of a-adrenoceptors is more appropriately based on pharmacological activity and additional subtypes of a-adrenoceptors have been identified by radioligand binding and molecular biological techniques [2]. -

Molecular Mechanisms Underlying Ketamine-Mediated Inhibition Of
Anesthesiology 2005; 102:93–101 © 2004 American Society of Anesthesiologists, Inc. Lippincott Williams & Wilkins, Inc. Molecular Mechanisms Underlying Ketamine-mediated Inhibition of Sarcolemmal Adenosine Triphosphate- sensitive Potassium Channels Takashi Kawano, M.D.,* Shuzo Oshita, M.D.,† Akira Takahashi, M.D.,‡ Yasuo Tsutsumi, M.D.,* Katsuya Tanaka, M.D.,§ Yoshinobu Tomiyama, M.D., Hiroshi Kitahata, M.D.,# Yutaka Nakaya, M.D.** Background: Ketamine inhibits adenosine triphosphate-sen- sensitive channel pore.3 These channels, as metabolic sitive potassium (KATP) channels, which results in the blocking sensors, are associated with such cellular functions as of ischemic preconditioning in the heart and inhibition of va- insulin secretion, cardiac preconditioning, vasodilata- sorelaxation induced by KATP channel openers. In the current 4–7 Downloaded from http://pubs.asahq.org/anesthesiology/article-pdf/102/1/93/357819/0000542-200501000-00017.pdf by guest on 27 September 2021 study, the authors investigated the molecular mechanisms of tion, and neuroprotection. ketamine’s actions on sarcolemmal KATP channels that are re- In cardiac myocytes, intravenous general anesthetics, associated by expressed subunits, inwardly rectifying potas- such as ketamine racemate, propofol, and thiamylal, di- sium channels (Kir6.1 or Kir6.2) and sulfonylurea receptors 8–10 rectly inhibit native sarcolemmal KATP channels. Al- (SUR1, SUR2A, or SUR2B). though these observations suggest that intravenous an- Methods: The authors used inside-out patch clamp configura- tions to investigate the effects of ketamine on the activities of esthetics may impair the endogenous organ protective reassociated Kir6.0/SUR channels containing wild-type, mutant, mechanisms mediated by KATP channels, the possibility or chimeric SURs expressed in COS-7 cells. -

Index Vol. 12-15
353 INDEX VOL. 12-15 Die Stichworte des Sachregisters sind in der jeweiligen Sprache der einzelnen Beitrage aufgefiihrt. Les termes repris dans la Table des matieres sont donnes selon la langue dans laquelle l'ouvrage est ecrit. The references of the Subject Index are given in the language of the respective contribution. 14 AAG (Alpha-acid glycoprotein) 120 14 Adenosine 108 12 Abortion 151 12 Adenosine-phosphate 311 13 Abscisin 12, 46, 66 13 Adenosine-5'-phosphosulfate 148 14 Absorbierbarkeit 317 13 Adenosine triphosphate 358 14 Absorption 309, 350 15 S-Adenosylmethionine 261 13 Absorption of drugs 139 13 Adipaenin (Spasmolytin) 318 14 - 15 12 Adrenal atrophy 96 14 Absorptionsgeschwindigkeit 300, 306 14 - 163, 164 14 Absorptionsquote 324 13 Adrenal gland 362 14 ACAI (Anticorticocatabolic activity in 12 Adrenalin(e) 319 dex) 145 14 - 209, 210 12 Acalo 197 15 - 161 13 Aceclidine (3-Acetoxyquinuclidine) 307, 13 {i-Adrenergic blockers 119 308, 310, 311, 330, 332 13 Adrenergic-blocking activity 56 13 Acedapsone 193,195,197 14 O(-Adrenergic blocking drugs 36, 37, 43 13 Aceperone (Acetabutone) 121 14 {i-Adrenergic blocking drugs 38 12 Acepromazin (Plegizil) 200 14 Adrenergic drugs 90 15 Acetanilid 156 12 Adrenocorticosteroids 14, 30 15 Acetazolamide 219 12 Adrenocorticotropic hormone (ACTH) 13 Acetoacetyl-coenzyme A 258 16,30,155 12 Acetohexamide 16 14 - 149,153,163,165,167,171 15 1-Acetoxy-8-aminooctahydroindolizin 15 Adrenocorticotropin (ACTH) 216 (Slaframin) 168 14 Adrenosterone 153 13 4-Acetoxy-1-azabicyclo(3, 2, 2)-nonane 12 Adreson 252 -

18512582.Pdf
European Heart Journal (2001) 22, 1938–1947 doi:10.1053/euhj.2001.2627, available online at http://www.idealibrary.com on The TRAPIST Study A multicentre randomized placebo controlled clinical trial of trapidil for prevention of restenosis after coronary stenting, measured by 3-D intravascular ultrasound P. W. Serruys1, D. P. Foley1, M. Pieper2, J. A. Kleijne3 and P. J. de Feyter1 on behalf of the TRAPIST investigators 1Department of Interventional Cardiology, Thoraxcenter, Erasmus Medical Center, Rotterdam, The Netherlands; 2Heartcenter Bodensee, Kreuzlingen, Switzerland; and 3Cardialysis, Rotterdam, The Netherlands Background Studies have reported benefit of oral therapy significant difference between trapidil and placebo-treated with the phosphodiesterase inhibitor, trapidil, in reducing patients regarding in-stent neointimal volume (108·6 restenosis after coronary angioplasty. Coronary stenting is 95·6 mm3 vs 93·379·1 mm3; P=0·16) or % obstruction associated with improved late outcome compared with volume (3818% vs 3621%; P=0·32), in angiographic balloon angioplasty, but significant neointimal hyperplasia minimal luminal diameter at follow-up (1·630·61 mm vs still occurs in a considerable proportion of patients. The 1·740·69 mm; P=0·17), restenosis rate (31% vs 24%; aim of this study was to investigate the safety and efficacy P=0·24), cumulative incidence of major adverse cardiac of trapidil 200 mg in preventing in-stent restenosis. events at 7 months (22% vs 20%; P=0·71) or anginal complaints (30% vs 24%; P=0·29). Methods Patients with a single native coronary lesion requiring revascularization were randomized to placebo or Conclusion Oral trapidil 600 mg daily for 6 months did trapidil at least 1 h before, and continuing for 6 months not reduce in-stent hyperplasia or improve clinical outcome after, successful implantation of a coronary Wallstent. -

(2006.01) Published: — A61K 9/00
ll ( (51) International Patent Classification: Published: A61K9/46 (2006.01) A61K /20 (2006.01) — with international search report (Art. 21(3)) A61K 9/00 (2006.01) A61P 1/04 (2006.01) A61K9/06 (2006.01) A61K 31/58 (2006.01) A61K 9/10 (2006.01) (21) International Application Number: PCT/EP20 19/0687 18 (22) International Filing Date: 11 July 2019 ( 11.07.2019) (25) Filing Language: English (26) Publication Language: English (30) Priority Data: 18182910.2 11 July 2018 ( 11.07.2018) EP (71) Applicant: MEDIZINISCHE UNIVERSITAT WIEN [AT/AT]; Spitalgasse 23, 1090 Wien (AT). (72) Inventors: ORGLER, Elisabeth; Leonhardstralk 3, 8010 Graz (AT). GASCHE, Christoph; Babenbergergasse 20, 3400 Klostemeuburg (AT). DABSCH, Stefanie; Alszeile 84, 1170 Wien (AT). (74) Agent: VOSSIUS & PARTNER (NO 31); Patentanwalte Rechtsanwalte mbB, Siebertstrasse 3, 81675 Munchen (PE). (81) Designated States (unless otherwise indicated, for every kind of national protection available) : AE, AG, AL, AM, AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, HR, HU, ID, IL, IN, IR, IS, JO, JP, KE, KG, KH, KN, KP, KR, KW, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. -

Iatrogenic Misadventure
BRITISH MEDICAL JOURNAL 18 MARcH 1972 733 Scientific Basis of Clinical Practice Br Med J: first published as 10.1136/bmj.1.5802.733 on 18 March 1972. Downloaded from Iatrogenic Misadventure A. G. W. WHITFIELD British Medical journal, 1972, 1, 733-734 effects. No one would pretend that our knowledge of drugs interacting with oral anticoagulants is complete. Even so, A striking feature of the history of therapeutics is the extent to sufficient is already known to indicate that anticoagulant drugs which fashion, rather than clinical science, governs the treat- have potent dangers and should be used only within the limited ment we mete out to our patients. Some ten years ago I attended sphere in which they are of proven worth. Moreover, when they a symposium on anticoagulant therapy and returned home are given, if possible, no other drug should be given at the firmly convinced that in all but a very few conditions such same time as many are known to inhibit or enhance their effect treatment was absolutely essential and indeed it was little short and many others may have similar interactions, even though of criminal negligence to withhold it. At that time the patient these are as yet unrecognized and unreported. queue for "prothrombin times" was usually the longest in our hospitals and phenindione was among the most frequently prescribed drugs, but our constant use of it stemmed from enthusiasm and ignorance of its dangers and not from scientific Hypotensive Drugs proof of its therapeutic advantages. Indeed it is only as know- Hypotensive drugs are being used increasingly and undoubtedly ledge has accumulated that we have learned of the toxic effects, their development has provided an enormous advance both in of the drug we gave so freely, on the kidney, the liver, the bone the treatment and in the prognosis of the severe forms of marrow, and the skin and that except in thromboembolic disease and in patients with hypertension-particularly in the younger age groups. -

(12) United States Patent (10) Patent No.: US 7.803,838 B2 Davis Et Al
USOO7803838B2 (12) United States Patent (10) Patent No.: US 7.803,838 B2 Davis et al. (45) Date of Patent: Sep. 28, 2010 (54) COMPOSITIONS COMPRISING NEBIVOLOL 2002fO169134 A1 11/2002 Davis 2002/0177586 A1 11/2002 Egan et al. (75) Inventors: Eric Davis, Morgantown, WV (US); 2002/0183305 A1 12/2002 Davis et al. John O'Donnell, Morgantown, WV 2002/0183317 A1 12/2002 Wagle et al. (US); Peter Bottini, Morgantown, WV 2002/0183365 A1 12/2002 Wagle et al. (US) 2002/0192203 A1 12, 2002 Cho 2003, OOO4194 A1 1, 2003 Gall (73) Assignee: Forest Laboratories Holdings Limited 2003, OO13699 A1 1/2003 Davis et al. (BM) 2003/0027820 A1 2, 2003 Gall (*) Notice: Subject to any disclaimer, the term of this 2003.0053981 A1 3/2003 Davis et al. patent is extended or adjusted under 35 2003, OO60489 A1 3/2003 Buckingham U.S.C. 154(b) by 455 days. 2003, OO69221 A1 4/2003 Kosoglou et al. 2003/0078190 A1* 4/2003 Weinberg ...................... 514f1 (21) Appl. No.: 11/141,235 2003/0078517 A1 4/2003 Kensey 2003/01 19428 A1 6/2003 Davis et al. (22) Filed: May 31, 2005 2003/01 19757 A1 6/2003 Davis 2003/01 19796 A1 6/2003 Strony (65) Prior Publication Data 2003.01.19808 A1 6/2003 LeBeaut et al. US 2005/027281.0 A1 Dec. 8, 2005 2003.01.19809 A1 6/2003 Davis 2003,0162824 A1 8, 2003 Krul Related U.S. Application Data 2003/0175344 A1 9, 2003 Waldet al. (60) Provisional application No. 60/577,423, filed on Jun. -

)&F1y3x PHARMACEUTICAL APPENDIX to THE
)&f1y3X PHARMACEUTICAL APPENDIX TO THE HARMONIZED TARIFF SCHEDULE )&f1y3X PHARMACEUTICAL APPENDIX TO THE TARIFF SCHEDULE 3 Table 1. This table enumerates products described by International Non-proprietary Names (INN) which shall be entered free of duty under general note 13 to the tariff schedule. The Chemical Abstracts Service (CAS) registry numbers also set forth in this table are included to assist in the identification of the products concerned. For purposes of the tariff schedule, any references to a product enumerated in this table includes such product by whatever name known. Product CAS No. Product CAS No. ABAMECTIN 65195-55-3 ACTODIGIN 36983-69-4 ABANOQUIL 90402-40-7 ADAFENOXATE 82168-26-1 ABCIXIMAB 143653-53-6 ADAMEXINE 54785-02-3 ABECARNIL 111841-85-1 ADAPALENE 106685-40-9 ABITESARTAN 137882-98-5 ADAPROLOL 101479-70-3 ABLUKAST 96566-25-5 ADATANSERIN 127266-56-2 ABUNIDAZOLE 91017-58-2 ADEFOVIR 106941-25-7 ACADESINE 2627-69-2 ADELMIDROL 1675-66-7 ACAMPROSATE 77337-76-9 ADEMETIONINE 17176-17-9 ACAPRAZINE 55485-20-6 ADENOSINE PHOSPHATE 61-19-8 ACARBOSE 56180-94-0 ADIBENDAN 100510-33-6 ACEBROCHOL 514-50-1 ADICILLIN 525-94-0 ACEBURIC ACID 26976-72-7 ADIMOLOL 78459-19-5 ACEBUTOLOL 37517-30-9 ADINAZOLAM 37115-32-5 ACECAINIDE 32795-44-1 ADIPHENINE 64-95-9 ACECARBROMAL 77-66-7 ADIPIODONE 606-17-7 ACECLIDINE 827-61-2 ADITEREN 56066-19-4 ACECLOFENAC 89796-99-6 ADITOPRIM 56066-63-8 ACEDAPSONE 77-46-3 ADOSOPINE 88124-26-9 ACEDIASULFONE SODIUM 127-60-6 ADOZELESIN 110314-48-2 ACEDOBEN 556-08-1 ADRAFINIL 63547-13-7 ACEFLURANOL 80595-73-9 ADRENALONE -

WO 2015/072852 Al 21 May 2015 (21.05.2015) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2015/072852 Al 21 May 2015 (21.05.2015) P O P C T (51) International Patent Classification: (81) Designated States (unless otherwise indicated, for every A61K 36/84 (2006.01) A61K 31/5513 (2006.01) kind of national protection available): AE, AG, AL, AM, A61K 31/045 (2006.01) A61P 31/22 (2006.01) AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, A61K 31/522 (2006.01) A61K 45/06 (2006.01) BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, (21) International Application Number: HN, HR, HU, ID, IL, IN, IR, IS, JP, KE, KG, KN, KP, KR, PCT/NL20 14/050780 KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, MG, (22) International Filing Date: MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, OM, 13 November 2014 (13.1 1.2014) PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, (25) Filing Language: English TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (26) Publication Language: English (84) Designated States (unless otherwise indicated, for every (30) Priority Data: kind of regional protection available): ARIPO (BW, GH, 61/903,430 13 November 2013 (13. 11.2013) US GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, ST, SZ, TZ, UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, (71) Applicant: RJG DEVELOPMENTS B.V. -

Role of Proaggregatory and Antiaggregatory Prostaglandins in Hemostasis
Role of proaggregatory and antiaggregatory prostaglandins in hemostasis. Studies with combined thromboxane synthase inhibition and thromboxane receptor antagonism. P Gresele, … , G Pieters, J Vermylen J Clin Invest. 1987;80(5):1435-1445. https://doi.org/10.1172/JCI113223. Research Article Thromboxane synthase inhibition can lead to two opposing effects: accumulation of proaggregatory cyclic endoperoxides and increased formation of antiaggregatory PGI2 and PGD2. The elimination of the effects of the cyclic endoperoxides by an endoperoxide-thromboxane A2 receptor antagonist should enhance the inhibition of hemostasis by thromboxane synthase blockers. We have carried out a series of double-blind, placebo-controlled, crossover studies in healthy volunteers to check if this hypothesis may be operative in vivo in man. In a first study, in 10 healthy male volunteers, the combined administration of the thromboxane receptor antagonist BM 13.177 and the thromboxane synthase inhibitor dazoxiben gave stronger inhibition of platelet aggregation and prolonged the bleeding time more than either drug alone. In a second study, in 10 different healthy male volunteers, complete inhibition of cyclooxygenase with indomethacin reduced the prolongation of the bleeding time by the combination BM 13.177 plus dazoxiben. In a third study, in five volunteers, selective cumulative inhibition of platelet TXA2 synthesis by low-dose aspirin inhibited platelet aggregation and prolonged the bleeding time less than the combination BM 13.177 plus dazoxiben. In vitro, in -

(12) United States Patent (10) Patent No.: US 6,264,917 B1 Klaveness Et Al
USOO6264,917B1 (12) United States Patent (10) Patent No.: US 6,264,917 B1 Klaveness et al. (45) Date of Patent: Jul. 24, 2001 (54) TARGETED ULTRASOUND CONTRAST 5,733,572 3/1998 Unger et al.. AGENTS 5,780,010 7/1998 Lanza et al. 5,846,517 12/1998 Unger .................................. 424/9.52 (75) Inventors: Jo Klaveness; Pál Rongved; Dagfinn 5,849,727 12/1998 Porter et al. ......................... 514/156 Lovhaug, all of Oslo (NO) 5,910,300 6/1999 Tournier et al. .................... 424/9.34 FOREIGN PATENT DOCUMENTS (73) Assignee: Nycomed Imaging AS, Oslo (NO) 2 145 SOS 4/1994 (CA). (*) Notice: Subject to any disclaimer, the term of this 19 626 530 1/1998 (DE). patent is extended or adjusted under 35 O 727 225 8/1996 (EP). U.S.C. 154(b) by 0 days. WO91/15244 10/1991 (WO). WO 93/20802 10/1993 (WO). WO 94/07539 4/1994 (WO). (21) Appl. No.: 08/958,993 WO 94/28873 12/1994 (WO). WO 94/28874 12/1994 (WO). (22) Filed: Oct. 28, 1997 WO95/03356 2/1995 (WO). WO95/03357 2/1995 (WO). Related U.S. Application Data WO95/07072 3/1995 (WO). (60) Provisional application No. 60/049.264, filed on Jun. 7, WO95/15118 6/1995 (WO). 1997, provisional application No. 60/049,265, filed on Jun. WO 96/39149 12/1996 (WO). 7, 1997, and provisional application No. 60/049.268, filed WO 96/40277 12/1996 (WO). on Jun. 7, 1997. WO 96/40285 12/1996 (WO). (30) Foreign Application Priority Data WO 96/41647 12/1996 (WO).