Taxonomy and Systematics of Quercus Subgenus Cyclobalanopsis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Book CMU 5(2)
CMU. Journal (2006) Vol. 5(2) 169 Survey and Herbarium Specimens of Medicinal Vascular Flora of Doi Suthep-Pui Somporn Putiyanan1* and J.F. Maxwell2 1 Department of Pharmaceutical Science, Faculty of Pharmacy, Chiang Mai University, Chiang Mai 50200, Thailand 2 Department of Biology, Faculty of Science, Chiang Mai University, Chiang Mai 50200, Thailand *Corresponding author. E-mail: [email protected] ABSTRACT The herbarium includes over 9,285 specimens from 238 families (270 fam. in the word) in medicinal plant herbarium, Faculty of Pharmacy, Chiang Mai University. From July 1987 to September 1991, a total of 2,044 species have been collected from Doi Suthep- Pui National Park, some of which are of considerable economic, medicinal and botanical interest. Vascular plants in this national park comprise of 193 of the 228 known families of vascular plants in Thailand, including a new family record for the flora of Thailand (Lardizabalaceae), eleven species new records for Thailand, three emended descriptions, two new combinations, and at least two species, with several others that are probably undescribed and new to science. The lowland, mostly disturbed forests up to 350-950 m. elevation, are of two deciduous facies, viz., dipterocarp-oak and mixed (former teak) forest. Elevations above this to the summit of Doi Suthep (c.1,620 m.) and Doi Pui (c.1,685 m.) are primary evergreen (monsoon) with some residual pine on some of the ridges. There is a distinct dry season (December-May) during which there are fires and many of the lowland species flower and fruit, many become leafless while in the evergreen areas, there is no specific flowering or fruiting season, that is, the phenologies of the plants in this habitat vary according to each species throughout the year. -

Oaks (Quercus Spp.): a Brief History
Publication WSFNR-20-25A April 2020 Oaks (Quercus spp.): A Brief History Dr. Kim D. Coder, Professor of Tree Biology & Health Care / University Hill Fellow University of Georgia Warnell School of Forestry & Natural Resources Quercus (oak) is the largest tree genus in temperate and sub-tropical areas of the Northern Hemisphere with an extensive distribution. (Denk et.al. 2010) Oaks are the most dominant trees of North America both in species number and biomass. (Hipp et.al. 2018) The three North America oak groups (white, red / black, and golden-cup) represent roughly 60% (~255) of the ~435 species within the Quercus genus worldwide. (Hipp et.al. 2018; McVay et.al. 2017a) Oak group development over time helped determine current species, and can suggest relationships which foster hybridization. The red / black and white oaks developed during a warm phase in global climate at high latitudes in what today is the boreal forest zone. From this northern location, both oak groups spread together southward across the continent splitting into a large eastern United States pathway, and much smaller western and far western paths. Both species groups spread into the eastern United States, then southward, and continued into Mexico and Central America as far as Columbia. (Hipp et.al. 2018) Today, Mexico is considered the world center of oak diversity. (Hipp et.al. 2018) Figure 1 shows genus, sub-genus and sections of Quercus (oak). History of Oak Species Groups Oaks developed under much different climates and environments than today. By examining how oaks developed and diversified into small, closely related groups, the native set of Georgia oak species can be better appreciated and understood in how they are related, share gene sets, or hybridize. -

Bulletin of Natural History ®
FLORI'IDA MUSEUM BULLETIN OF NATURAL HISTORY ® A MIDDLE EOCENE FOSSIL PLANT ASSEMBLAGE (POWERS CLAY PIT) FROM WESTERN TENNESSEE DavidL. Dilcher and Terry A. Lott Vol. 45, No. 1, pp. 1-43 2005 UNIVERSITY OF FLORIDA GAINESVILLE - The FLORIDA MUSEUM OF NATURAL HiSTORY is Florida«'s state museum of natural history, dedicated to understanding, preser¥ingrand interpreting].biologica[1 diversity and culturafheritage. The BULLETIN OF THE FLORIDA- MUSEUM OF NATURAL HISTORY is a peer-reviewed publication thatpziblishes.the result5 of origifial reseafchin zodlogy, botany, paleontology, and archaeology. Address all inquiries t6 the Managing Editor ofthe Bulletin. Numbers,ofthe Bulletin,afe,published,at itregular intervals. Specific volumes are not'necessarily completed in anyone year. The end of a volume willl·be noted at the foot of the first page ofthe last issue in that volume. Richard Franz, Managing Editor Erika H. Simons, Production BulletinCommittee Richard Franz,,Chairperson Ann Cordell Sarah Fazenbaker Richard Hulbert WilliamMarquardt Susan Milbrath Irvy R. Quitmyer - Scott Robinson, Ex 01#cio Afember ISSN: 0071-6154 Publication Date: October 31,2005 Send communications concerning purchase or exchange of the publication and manustfipt queries to: Managing Editor of the BULLETIN Florida MuseumofNatural-History University offlorida PO Box 117800 Gainesville, FL 32611 -7800 U.S.A. Phone: 352-392-1721 Fax: 352-846-0287 e-mail: [email protected] A MIDDLE EOCENE FOSSIL PLANT ASSEMBLAGE (POWERS CLAY PIT) FROM WESTERN TENNESSEE David L. Dilcher and Terry A. Lottl ABSTRACT Plant megafossils are described, illustrated and discussed from Powers Clay Pit, occurring in the middle Eocene, Claiborne Group of the Mississippi Embayment in western Tennessee. -

University of California Santa Cruz Responding to An
UNIVERSITY OF CALIFORNIA SANTA CRUZ RESPONDING TO AN EMERGENT PLANT PEST-PATHOGEN COMPLEX ACROSS SOCIAL-ECOLOGICAL SCALES A dissertation submitted in partial satisfaction of the requirements for the degree of DOCTOR OF PHILOSOPHY in ENVIRONMENTAL STUDIES with an emphasis in ECOLOGY AND EVOLUTIONARY BIOLOGY by Shannon Colleen Lynch December 2020 The Dissertation of Shannon Colleen Lynch is approved: Professor Gregory S. Gilbert, chair Professor Stacy M. Philpott Professor Andrew Szasz Professor Ingrid M. Parker Quentin Williams Acting Vice Provost and Dean of Graduate Studies Copyright © by Shannon Colleen Lynch 2020 TABLE OF CONTENTS List of Tables iv List of Figures vii Abstract x Dedication xiii Acknowledgements xiv Chapter 1 – Introduction 1 References 10 Chapter 2 – Host Evolutionary Relationships Explain 12 Tree Mortality Caused by a Generalist Pest– Pathogen Complex References 38 Chapter 3 – Microbiome Variation Across a 66 Phylogeographic Range of Tree Hosts Affected by an Emergent Pest–Pathogen Complex References 110 Chapter 4 – On Collaborative Governance: Building Consensus on 180 Priorities to Manage Invasive Species Through Collective Action References 243 iii LIST OF TABLES Chapter 2 Table I Insect vectors and corresponding fungal pathogens causing 47 Fusarium dieback on tree hosts in California, Israel, and South Africa. Table II Phylogenetic signal for each host type measured by D statistic. 48 Table SI Native range and infested distribution of tree and shrub FD- 49 ISHB host species. Chapter 3 Table I Study site attributes. 124 Table II Mean and median richness of microbiota in wood samples 128 collected from FD-ISHB host trees. Table III Fungal endophyte-Fusarium in vitro interaction outcomes. -

16Th Annual NECLIME Meeting ABSTRACTS
16th Annual NECLIME Meeting Madrid, October 14 – 17, 2015 ABSTRACTS 16th NECLIME Meeting Madrid, October 14–17, 2015 16th Annual NECLIME Meeting Geominero Museum Geological Survey of Spain (Instituto Geológico y Minero de España - IGME) Madrid – October 14–17, 2015 Under the sponsorship of the Department of Geology, Faculty of Sciences, University of Salamanca and the Research Project nº CGL2011-23438/BTE (Environmental characterization of Miocene lacustrine systems with marine-like faunas from the Duero and Ebro basins: geochemistry of biogenic carbonates and palynology), Instituto de Ciencias de la Tierra Jaume Almera (Spanish Council for Scientific Research - CSIC). ABSTRACTS Eduardo Barrón (Ed.) 3 16th NECLIME Meeting Madrid, October 14–17, 2015 ORGANIZING COMMITTEE Chairman: - María F. Valle, Salamanca University, Spain Executive Secretary: - Eduardo Barrón, Geological Survey of Spain, Madrid Members: - Angela A. Bruch, Senckenberg Research Institute, Frankfurt am Main, Germany - Manuel Casas-Gallego, Robertson (UK) Ltd., United Kingdom - José María Postigo-Mijarra, School of Forestry Engineering. Technical University of Madrid - Isabel Rábano Gutiérrez del Arroyo, Geological Survey of Spain, Madrid - Mª Rosario Rivas-Carballo, Salamanca University, Spain - Torsten Utescher, Steinmann Institute, Bonn University, Germany 4 16th NECLIME Meeting Madrid, October 14–17, 2015 PROGRAMME Wednesday, October 14th Geominero Museum (Instituto Geológico y Minero de España, IGME) 16.00-18.00 Reception of participants 18.00-19.00 Guided visit to the Museum 19.00... Short walking city tour through the centre of Madrid Thursday Morning, October 15th 9.30-10.00 Reception of participants 10.00-10.15 Inauguration of 16th NECLIME Meeting 10.15-10.45 Introduction to NECLIME and information about the latest activities 10.45-11.45 Invited conference: Reconstructing palaeofloras based on fossils, climate and phylogenies Dr. -

Quercus ×Coutinhoi Samp. Discovered in Australia Charlie Buttigieg
XXX International Oaks The Journal of the International Oak Society …the hybrid oak that time forgot, oak-rod baskets, pros and cons of grafting… Issue No. 25/ 2014 / ISSN 1941-2061 1 International Oaks The Journal of the International Oak Society … the hybrid oak that time forgot, oak-rod baskets, pros and cons of grafting… Issue No. 25/ 2014 / ISSN 1941-2061 International Oak Society Officers and Board of Directors 2012-2015 Officers President Béatrice Chassé (France) Vice-President Charles Snyers d’Attenhoven (Belgium) Secretary Gert Fortgens (The Netherlands) Treasurer James E. Hitz (USA) Board of Directors Editorial Committee Membership Director Chairman Emily Griswold (USA) Béatrice Chassé Tour Director Members Shaun Haddock (France) Roderick Cameron International Oaks Allen Coombes Editor Béatrice Chassé Shaun Haddock Co-Editor Allen Coombes (Mexico) Eike Jablonski (Luxemburg) Oak News & Notes Ryan Russell Editor Ryan Russell (USA) Charles Snyers d’Attenhoven International Editor Roderick Cameron (Uruguay) Website Administrator Charles Snyers d’Attenhoven For contributions to International Oaks contact Béatrice Chassé [email protected] or [email protected] 0033553621353 Les Pouyouleix 24800 St.-Jory-de-Chalais France Author’s guidelines for submissions can be found at http://www.internationaloaksociety.org/content/author-guidelines-journal-ios © 2014 International Oak Society Text, figures, and photographs © of individual authors and photographers. Graphic design: Marie-Paule Thuaud / www.lecentrecreatifducoin.com Photos. Cover: Charles Snyers d’Attenhoven (Quercus macrocalyx Hickel & A. Camus); p. 6: Charles Snyers d’Attenhoven (Q. oxyodon Miq.); p. 7: Béatrice Chassé (Q. acerifolia (E.J. Palmer) Stoynoff & W. J. Hess); p. 9: Eike Jablonski (Q. ithaburensis subsp. -
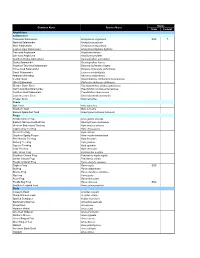
St. Joseph Bay Native Species List
Status Common Name Species Name State Federal Amphibians Salamanders Flatwoods Salamander Ambystoma cingulatum SSC T Marbled Salamander Ambystoma opacum Mole Salamander Ambystoma talpoideum Eastern Tiger Salamander Ambystoma tigrinum tigrinum Two-toed Amphiuma Amphiuma means One-toed Amphiuma Amphiuma pholeter Southern Dusky Salamander Desmognathus auriculatus Dusky Salamander Desmognathus fuscus Southern Two-lined Salamander Eurycea bislineata cirrigera Three-lined Salamander Eurycea longicauda guttolineata Dwarf Salamander Eurycea quadridigitata Alabama Waterdog Necturus alabamensis Central Newt Notophthalmus viridescens louisianensis Slimy Salamander Plethodon glutinosus glutinosus Slender Dwarf Siren Pseudobranchus striatus spheniscus Gulf Coast Mud Salamander Pseudotriton montanus flavissimus Southern Red Salamander Pseudotriton ruber vioscai Eastern Lesser Siren Siren intermedia intermedia Greater Siren Siren lacertina Toads Oak Toad Bufo quercicus Southern Toad Bufo terrestris Eastern Spadefoot Toad Scaphiopus holbrooki holbrooki Frogs Florida Cricket Frog Acris gryllus dorsalis Eastern Narrow-mouthed Frog Gastrophryne carolinensis Western Bird-voiced Treefrog Hyla avivoca avivoca Cope's Gray Treefrog Hyla chrysoscelis Green Treefrog Hyla cinerea Southern Spring Peeper Hyla crucifer bartramiana Pine Woods Treefrog Hyla femoralis Barking Treefrog Hyla gratiosa Squirrel Treefrog Hyla squirella Gray Treefrog Hyla versicolor Little Grass Frog Limnaoedus ocularis Southern Chorus Frog Pseudacris nigrita nigrita Ornate Chorus Frog Pseudacris -

The Effect of Season of Fire on the Recovery of Florida Scrub
2B.7 THE EFFECT OF SEASON OF FIRE ON THE RECOVERY OF FLORIDA SCRUB Tammy E. Foster* and Paul A. Schmalzer Dynamac Corporation, Kennedy Space Center, Florida 1. ABSTRACT∗ scrub, rosemary scrub, and scrubby flatwoods (Myers 1990, Abrahamson and Hartnett 1990). Florida scrub is a xeromorphic shrubland that is Florida scrub is habitat for threatened and maintained by frequent fires. Historically, these fires endangered plants and animals (Christman and Judd occurred during the summer due to lightning ignition. 1990, Stout and Marion 1993, Stout 2001). Today, Florida scrub is often managed by the use of Management of remaining scrub is critical to the survival prescribed burning. Prescribed burning of scrub has of these species. Scrub is a fire-maintained system been implemented on Kennedy Space Center/Merritt (Myers 1990, Menges 1999), and recovery after fire of Island National Wildlife Refuge (KSC/MINWR) since oak scrub and scrubby flatwoods is primarily through 1981, with burns being carried out throughout the year. sprouting and, in some species, clonal spread of the The impacts of the season of burn on recovery are not dominant shrubs (Abrahamson 1984a, 1984b, known. Long-term monitoring of scrub regeneration has Schmalzer and Hinkle 1992a, Menges and Hawkes been conducted since the early-1980’s at KSC/MINWR 1998). Scrub naturally burned during the summer using permanent 15 m line-intercept transects. We months due to lightning ignition. However, landscape obtained data from eight transects that were subjected fragmentation and fire suppression have reduced fire to a winter burn in 1986 and a summer burn in 1997 and size and frequency (Myers 1990, Duncan and compared the recovery of the stand for the first five Schmalzer 2001). -

De Novo Transcriptome Analysis of Dalbergia Odorifera T
Article De Novo Transcriptome Analysis of Dalbergia odorifera T. Chen (Fabaceae) and Transferability of SSR Markers Developed from the Transcriptome Fu-Mei Liu 1,2 , Zhou Hong 3,*, Zeng-Jiang Yang 3, Ning-Nan Zhang 3, Xiao-Jin Liu 3 and Da-Ping Xu 3 1 State Key Laboratory of Tree Genetics and Breeding, Northeast Forestry University, Harbin 150040, China; [email protected] 2 The Experimental Centre of Tropical Forestry, Chinese Academy of Forestry, Pingxiang 532600, China 3 Research Institute of Tropical Forestry, Chinese Academy of Forestry, Longdong, Guangzhou 510520, China; [email protected] (Z.-J.Y.); [email protected] (N.-N.Z.); [email protected] (X.-J.L.); [email protected] (D.-P.X.) * Correspondence: [email protected]; Tel.: +86-20-8703-1037 Received: 15 December 2018; Accepted: 24 January 2019; Published: 26 January 2019 Abstract: Dalbergia odorifera T. Chen (Fabaceae), indigenous to Hainan Island, is a precious rosewood (Hainan hualimu) in China. However, only limited genomic information is available which has resulted in a lack of molecular markers, limiting the development and utilization of the germplasm resources. In this study, we aim to enrich genomic information of D. odorifera, and develop a series of transferable simple sequence repeat (SSR) markers for Dalbergia species. Therefore, we performed transcriptome sequencing for D. odorifera by pooling leaf tissues from three trees. A dataset of 138,516,418 reads was identified and assembled into 115,292 unigenes. Moreover, 35,774 simple sequence repeats (SSRs) were identified as potential SSR markers. A set of 19 SSR markers was successfully transferred across species of Dalbergia odorifera T. -

Assessing Restoration Potential of Fragmented and Degraded Fagaceae Forests in Meghalaya, North-East India
Article Assessing Restoration Potential of Fragmented and Degraded Fagaceae Forests in Meghalaya, North-East India Prem Prakash Singh 1,2,* , Tamalika Chakraborty 3, Anna Dermann 4 , Florian Dermann 4, Dibyendu Adhikari 1, Purna B. Gurung 1, Saroj Kanta Barik 1,2, Jürgen Bauhus 4 , Fabian Ewald Fassnacht 5, Daniel C. Dey 6, Christine Rösch 7 and Somidh Saha 4,7,* 1 Department of Botany, North-Eastern Hill University, Shillong 793022, India; [email protected] (D.A.); [email protected] (P.B.G.); [email protected] (S.K.B.) 2 CSIR-National Botanical Research Institute, Council of Scientific & Industrial Research, Rana Pratap Marg, Lucknow 226001, Uttar Pradesh, India 3 Institute of Forest Ecosystems, Thünen Institute, Alfred-Möller-Str. 1, House number 41/42, D-16225 Eberswalde, Germany; [email protected] 4 Chair of Silviculture, University of Freiburg, Tennenbacherstr. 4, D-79085 Freiburg, Germany; anna-fl[email protected] (A.D.); fl[email protected] (F.D.); [email protected] (J.B.) 5 Institute for Geography and Geoecology, Karlsruhe Institute of Technology, Kaiserstr. 12, D-76131 Karlsruhe, Germany; [email protected] 6 Northern Research Station, USDA Forest Service, 202 Natural Resources Building, Columbia, MO 65211-7260, USA; [email protected] 7 Institute for Technology Assessment and Systems Analysis, Karlsruhe Institute of Technology, Karlstr. 11, D-76133 Karlsruhe, Germany; [email protected] * Correspondence: prem12fl[email protected] (P.P.S.); [email protected] (S.S.) Received: 5 August 2020; Accepted: 16 September 2020; Published: 19 September 2020 Abstract: The montane subtropical broad-leaved humid forests of Meghalaya (Northeast India) are highly diverse and situated at the transition zone between the Eastern Himalayas and Indo-Burma biodiversity hotspots. -

An Evolutionary Perspective on Human Cross-Sensitivity to Tree Nut and Seed Allergens," Aliso: a Journal of Systematic and Evolutionary Botany: Vol
Aliso: A Journal of Systematic and Evolutionary Botany Volume 33 | Issue 2 Article 3 2015 An Evolutionary Perspective on Human Cross- sensitivity to Tree Nut and Seed Allergens Amanda E. Fisher Rancho Santa Ana Botanic Garden, Claremont, California, [email protected] Annalise M. Nawrocki Pomona College, Claremont, California, [email protected] Follow this and additional works at: http://scholarship.claremont.edu/aliso Part of the Botany Commons, Evolution Commons, and the Nutrition Commons Recommended Citation Fisher, Amanda E. and Nawrocki, Annalise M. (2015) "An Evolutionary Perspective on Human Cross-sensitivity to Tree Nut and Seed Allergens," Aliso: A Journal of Systematic and Evolutionary Botany: Vol. 33: Iss. 2, Article 3. Available at: http://scholarship.claremont.edu/aliso/vol33/iss2/3 Aliso, 33(2), pp. 91–110 ISSN 0065-6275 (print), 2327-2929 (online) AN EVOLUTIONARY PERSPECTIVE ON HUMAN CROSS-SENSITIVITY TO TREE NUT AND SEED ALLERGENS AMANDA E. FISHER1-3 AND ANNALISE M. NAWROCKI2 1Rancho Santa Ana Botanic Garden and Claremont Graduate University, 1500 North College Avenue, Claremont, California 91711 (Current affiliation: Department of Biological Sciences, California State University, Long Beach, 1250 Bellflower Boulevard, Long Beach, California 90840); 2Pomona College, 333 North College Way, Claremont, California 91711 (Current affiliation: Amgen Inc., [email protected]) 3Corresponding author ([email protected]) ABSTRACT Tree nut allergies are some of the most common and serious allergies in the United States. Patients who are sensitive to nuts or to seeds commonly called nuts are advised to avoid consuming a variety of different species, even though these may be distantly related in terms of their evolutionary history. -

Northern Yellow Bat Roost Selection and Fidelity in South Carolina
Final Report South Carolina State Wildlife Grant SC T-F16AF00598 South Carolina Department of Natural Resources (SCDNR) May 1, 2016-June 30, 2017 Project Title: Northern Yellow Bat Roost Selection and Fidelity in South Carolina Mary Socci, Palmetto Bluff Conservancy (PBC), Jay Walea, PBC, Timothy White, PBC, Jason Robinson, Biological Systems Consultants, Inc., and Jennifer Kindel, SCDNR Objective 1: Radio-track healthy Northern yellow bats (≤ 10) captured by mist netting appropriate habitat in spring, summer, and fall 2016, ideally with at least 3 radio-tracking events in each season. Record roost switching, and describe roost sites selected. Accomplishments: Introduction: The primary purpose of this study was to investigate the roost site selection and fidelity of northern yellow bats (Lasiurus intermedius, syn. Dasypterus intermedius) by capturing, radio- tagging and tracking individual L. intermedius at Palmetto Bluff, a 15,000 acre, partially-developed tract in Beaufort County, South Carolina (Figures 1 and 2). Other objectives (2 and 3) were to obtain audio recordings of bats foraging in various habitats across the Palmetto Bluff property, including as many L. intermedius as possible and to initiate a public outreach program in order to educate the community on both the project and the environmental needs of bats, many of which are swiftly declining species in United States. Figure 1. Location of Palmetto Bluff 1 Figure 2. The Palmetto Bluff Development Tract The life history of northern yellow bats, a high-priority species in the Southeast, is poorly understood. Studies in coastal Georgia found that all yellow bats that were tracked roosted in Spanish moss in southern live oaks (Quercus virginiana) and sand live oaks (Quercus geminata) (Coleman et al.