Table of Contents
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
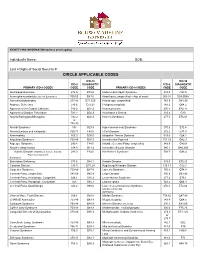
Circle Applicable Codes
IDENTIFYING INFORMATION (please print legibly) Individual’s Name: DOB: Last 4 Digits of Social Security #: CIRCLE APPLICABLE CODES ICD-10 ICD-10 ICD-9 DIAGNOSTIC ICD-9 DIAGNOSTIC PRIMARY ICD-9 CODES CODE CODE PRIMARY ICD-9 CODES CODE CODE Abetalipoproteinemia 272.5 E78.6 Hallervorden-Spatz Syndrome 333.0 G23.0 Acrocephalosyndactyly (Apert’s Syndrome) 755.55 Q87.0 Head Injury, unspecified – Age of onset: 959.01 S09.90XA Adrenaleukodystrophy 277.86 E71.529 Hemiplegia, unspecified 342.9 G81.90 Arginase Deficiency 270.6 E72.21 Holoprosencephaly 742.2 Q04.2 Agenesis of the Corpus Callosum 742.2 Q04.3 Homocystinuria 270.4 E72.11 Agenesis of Septum Pellucidum 742.2 Q04.3 Huntington’s Chorea 333.4 G10 Argyria/Pachygyria/Microgyria 742.2 Q04.3 Hurler’s Syndrome 277.5 E76.01 or 758.33 Aicardi Syndrome 333 G23.8 Hyperammonemia Syndrome 270.6 E72.4 Alcohol Embryo and Fetopathy 760.71 F84.5 I-Cell Disease 272.2 E77.0 Anencephaly 655.0 Q00.0 Idiopathic Torsion Dystonia 333.6 G24.1 Angelman Syndrome 759.89 Q93.5 Incontinentia Pigmenti 757.33 Q82.3 Asperger Syndrome 299.8 F84.5 Infantile Cerebral Palsy, unspecified 343.9 G80.9 Ataxia-Telangiectasia 334.8 G11.3 Intractable Seizure Disorder 345.1 G40.309 Autistic Disorder (Childhood Autism, Infantile 299.0 F84.0 Klinefelter’s Syndrome 758.7 Q98.4 Psychosis, Kanner’s Syndrome) Biotinidase Deficiency 277.6 D84.1 Krabbe Disease 333.0 E75.23 Canavan Disease 330.0 E75.29 Kugelberg-Welander Disease 335.11 G12.1 Carpenter Syndrome 759.89 Q87.0 Larsen’s Syndrome 755.8 Q74.8 Cerebral Palsy, unspecified 343.69 G80.9 -

Level Estimates of Maternal Smoking and Nicotine Replacement Therapy During Pregnancy
Using primary care data to assess population- level estimates of maternal smoking and nicotine replacement therapy during pregnancy Nafeesa Nooruddin Dhalwani BSc MSc Thesis submitted to the University of Nottingham for the degree of Doctor of Philosophy November 2014 ABSTRACT Background: Smoking in pregnancy is the most significant preventable cause of poor health outcomes for women and their babies and, therefore, is a major public health concern. In the UK there is a wide range of interventions and support for pregnant women who want to quit. One of these is nicotine replacement therapy (NRT) which has been widely available for retail purchase and prescribing to pregnant women since 2005. However, measures of NRT prescribing in pregnant women are scarce. These measures are vital to assess its usefulness in smoking cessation during pregnancy at a population level. Furthermore, evidence of NRT safety in pregnancy for the mother and child’s health so far is nebulous, with existing studies being small or using retrospectively reported exposures. Aims and Objectives: The main aim of this work was to assess population- level estimates of maternal smoking and NRT prescribing in pregnancy and the safety of NRT for both the mother and the child in the UK. Currently, the only population-level data on UK maternal smoking are from repeated cross-sectional surveys or routinely collected maternity data during pregnancy or at delivery. These obtain information at one point in time, and there are no population-level data on NRT use available. As a novel approach, therefore, this thesis used the routinely collected primary care data that are currently available for approximately 6% of the UK population and provide longitudinal/prospectively recorded information throughout pregnancy. -

MECHANISMS in ENDOCRINOLOGY: Novel Genetic Causes of Short Stature
J M Wit and others Genetics of short stature 174:4 R145–R173 Review MECHANISMS IN ENDOCRINOLOGY Novel genetic causes of short stature 1 1 2 2 Jan M Wit , Wilma Oostdijk , Monique Losekoot , Hermine A van Duyvenvoorde , Correspondence Claudia A L Ruivenkamp2 and Sarina G Kant2 should be addressed to J M Wit Departments of 1Paediatrics and 2Clinical Genetics, Leiden University Medical Center, PO Box 9600, 2300 RC Leiden, Email The Netherlands [email protected] Abstract The fast technological development, particularly single nucleotide polymorphism array, array-comparative genomic hybridization, and whole exome sequencing, has led to the discovery of many novel genetic causes of growth failure. In this review we discuss a selection of these, according to a diagnostic classification centred on the epiphyseal growth plate. We successively discuss disorders in hormone signalling, paracrine factors, matrix molecules, intracellular pathways, and fundamental cellular processes, followed by chromosomal aberrations including copy number variants (CNVs) and imprinting disorders associated with short stature. Many novel causes of GH deficiency (GHD) as part of combined pituitary hormone deficiency have been uncovered. The most frequent genetic causes of isolated GHD are GH1 and GHRHR defects, but several novel causes have recently been found, such as GHSR, RNPC3, and IFT172 mutations. Besides well-defined causes of GH insensitivity (GHR, STAT5B, IGFALS, IGF1 defects), disorders of NFkB signalling, STAT3 and IGF2 have recently been discovered. Heterozygous IGF1R defects are a relatively frequent cause of prenatal and postnatal growth retardation. TRHA mutations cause a syndromic form of short stature with elevated T3/T4 ratio. Disorders of signalling of various paracrine factors (FGFs, BMPs, WNTs, PTHrP/IHH, and CNP/NPR2) or genetic defects affecting cartilage extracellular matrix usually cause disproportionate short stature. -

Soonerstart Automatic Qualifying Syndromes and Conditions
SoonerStart Automatic Qualifying Syndromes and Conditions - Appendix O Abetalipoproteinemia Acanthocytosis (see Abetalipoproteinemia) Accutane, Fetal Effects of (see Fetal Retinoid Syndrome) Acidemia, 2-Oxoglutaric Acidemia, Glutaric I Acidemia, Isovaleric Acidemia, Methylmalonic Acidemia, Propionic Aciduria, 3-Methylglutaconic Type II Aciduria, Argininosuccinic Acoustic-Cervico-Oculo Syndrome (see Cervico-Oculo-Acoustic Syndrome) Acrocephalopolysyndactyly Type II Acrocephalosyndactyly Type I Acrodysostosis Acrofacial Dysostosis, Nager Type Adams-Oliver Syndrome (see Limb and Scalp Defects, Adams-Oliver Type) Adrenoleukodystrophy, Neonatal (see Cerebro-Hepato-Renal Syndrome) Aglossia Congenita (see Hypoglossia-Hypodactylia) Aicardi Syndrome AIDS Infection (see Fetal Acquired Immune Deficiency Syndrome) Alaninuria (see Pyruvate Dehydrogenase Deficiency) Albers-Schonberg Disease (see Osteopetrosis, Malignant Recessive) Albinism, Ocular (includes Autosomal Recessive Type) Albinism, Oculocutaneous, Brown Type (Type IV) Albinism, Oculocutaneous, Tyrosinase Negative (Type IA) Albinism, Oculocutaneous, Tyrosinase Positive (Type II) Albinism, Oculocutaneous, Yellow Mutant (Type IB) Albinism-Black Locks-Deafness Albright Hereditary Osteodystrophy (see Parathyroid Hormone Resistance) Alexander Disease Alopecia - Mental Retardation Alpers Disease Alpha 1,4 - Glucosidase Deficiency (see Glycogenosis, Type IIA) Alpha-L-Fucosidase Deficiency (see Fucosidosis) Alport Syndrome (see Nephritis-Deafness, Hereditary Type) Amaurosis (see Blindness) Amaurosis -

(12) Patent Application Publication (10) Pub. No.: US 2010/0210567 A1 Bevec (43) Pub
US 2010O2.10567A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2010/0210567 A1 Bevec (43) Pub. Date: Aug. 19, 2010 (54) USE OF ATUFTSINASATHERAPEUTIC Publication Classification AGENT (51) Int. Cl. A638/07 (2006.01) (76) Inventor: Dorian Bevec, Germering (DE) C07K 5/103 (2006.01) A6IP35/00 (2006.01) Correspondence Address: A6IPL/I6 (2006.01) WINSTEAD PC A6IP3L/20 (2006.01) i. 2O1 US (52) U.S. Cl. ........................................... 514/18: 530/330 9 (US) (57) ABSTRACT (21) Appl. No.: 12/677,311 The present invention is directed to the use of the peptide compound Thr-Lys-Pro-Arg-OH as a therapeutic agent for (22) PCT Filed: Sep. 9, 2008 the prophylaxis and/or treatment of cancer, autoimmune dis eases, fibrotic diseases, inflammatory diseases, neurodegen (86). PCT No.: PCT/EP2008/007470 erative diseases, infectious diseases, lung diseases, heart and vascular diseases and metabolic diseases. Moreover the S371 (c)(1), present invention relates to pharmaceutical compositions (2), (4) Date: Mar. 10, 2010 preferably inform of a lyophilisate or liquid buffersolution or artificial mother milk formulation or mother milk substitute (30) Foreign Application Priority Data containing the peptide Thr-Lys-Pro-Arg-OH optionally together with at least one pharmaceutically acceptable car Sep. 11, 2007 (EP) .................................. O7017754.8 rier, cryoprotectant, lyoprotectant, excipient and/or diluent. US 2010/0210567 A1 Aug. 19, 2010 USE OF ATUFTSNASATHERAPEUTIC ment of Hepatitis BVirus infection, diseases caused by Hepa AGENT titis B Virus infection, acute hepatitis, chronic hepatitis, full minant liver failure, liver cirrhosis, cancer associated with Hepatitis B Virus infection. 0001. The present invention is directed to the use of the Cancer, Tumors, Proliferative Diseases, Malignancies and peptide compound Thr-Lys-Pro-Arg-OH (Tuftsin) as a thera their Metastases peutic agent for the prophylaxis and/or treatment of cancer, 0008. -

Craniosynostosis Precision Panel Overview Indications Clinical Utility
Craniosynostosis Precision Panel Overview Craniosynostosis is defined as the premature fusion of one or more cranial sutures, often resulting in abnormal head shape. It is a developmental craniofacial anomaly resulting from a primary defect of ossification (primary craniosynostosis) or, more commonly, from a failure of brain growth (secondary craniosynostosis). As well, craniosynostosis can be simple when only one suture fuses prematurely or complex/compound when there is a premature fusion of multiple sutures. Complex craniosynostosis are usually associated with other body deformities. The main morbidity risk is the elevated intracranial pressure and subsequent brain damage. When left untreated, craniosynostosis can cause serious complications such as developmental delay, facial abnormality, sensory, respiratory and neurological dysfunction, eye anomalies and psychosocial disturbances. In approximately 85% of the cases, this disease is isolated and nonsyndromic. Syndromic craniosynostosis usually present with multiorgan complications. The Igenomix Craniosynostosis Precision Panel can be used to make a directed and accurate diagnosis ultimately leading to a better management and prognosis of the disease. It provides a comprehensive analysis of the genes involved in this disease using next-generation sequencing (NGS) to fully understand the spectrum of relevant genes involved. Indications The Igenomix Craniosynostosis Precision Panel is indicated for those patients with a clinical diagnosis or suspicion with or without the following manifestations: ‐ Microcephaly ‐ Scaphocephaly (elongated head) ‐ Anterior plagiocephaly ‐ Brachycephaly ‐ Torticollis ‐ Frontal bossing Clinical Utility The clinical utility of this panel is: - The genetic and molecular confirmation for an accurate clinical diagnosis of a symptomatic patient. - Early initiation of treatment in the form surgical procedures to relieve fused sutures, midface advancement, limited phase of orthodontic treatment and combined 1 orthodontics/orthognathic surgery treatment. -

Early ACCESS Diagnosed Conditions List
Iowa Early ACCESS Diagnosed Conditions Eligibility List List adapted with permission from Early Intervention Colorado To search for a specific word type "Ctrl F" to use the "Find" function. Is this diagnosis automatically eligible for Early Medical Diagnosis Name Other Names for the Diagnosis and Additional Diagnosis Information ACCESS? 6q terminal deletion syndrome Yes Achondrogenesis I Parenti-Fraccaro Yes Achondrogenesis II Langer-Saldino Yes Schinzel Acrocallosal syndrome; ACLS; ACS; Hallux duplication, postaxial polydactyly, and absence of the corpus Acrocallosal syndrome, Schinzel Type callosum Yes Acrodysplasia; Arkless-Graham syndrome; Maroteaux-Malamut syndrome; Nasal hypoplasia-peripheral dysostosis-intellectual disability syndrome; Peripheral dysostosis-nasal hypoplasia-intellectual disability (PNM) Acrodysostosis syndrome Yes ALD; AMN; X-ALD; Addison disease and cerebral sclerosis; Adrenomyeloneuropathy; Siemerling-creutzfeldt disease; Bronze schilder disease; Schilder disease; Melanodermic Leukodystrophy; sudanophilic leukodystrophy; Adrenoleukodystrophy Pelizaeus-Merzbacher disease Yes Agenesis of Corpus Callosum Absence of the corpus callosum; Hypogenesis of the corpus callosum; Dysplastic corpus callosum Yes Agenesis of Corpus Callosum and Chorioretinal Abnormality; Agenesis of Corpus Callosum With Chorioretinitis Abnormality; Agenesis of Corpus Callosum With Infantile Spasms And Ocular Anomalies; Chorioretinal Anomalies Aicardi syndrome with Agenesis Yes Alexander Disease Yes Allan Herndon syndrome Allan-Herndon-Dudley -

Back Matter 611-642.Pdf
Index A Acrodysostosis, 527, 538 Aarskog syndrome, 527, 537 Acrofacial dysostosis 1, 580–581 differentiated from Opitz G/BBB Acrofacial dysplasia, 563 syndrome, 458 Acro-fronto-facio-nasal dysostosis Aase-Smith syndrome, 527 syndrome, 527, 538–539 Aase syndrome, 527, 537 Acromegaloid facial appearance Abducens nerve. See Cranial nerve VI syndrome, 527, 539 Abduction, Möbius sequence-related Acromegaloid phenotype-cutis verticis impairment of, 197, 198 gyrata-cornea leukoma, 527, 539 Abetalipoproteinemia, 527, 537 Acromesomelic dysplasia, Maroteaux- Ablepharon-macrostomia syndrome, Martinelli-Campailla type, 527, 527, 538 539 Abortion, spontaneous, chromosomal Acromicric dysplasia, 527, 539 abnormalities associated with, 83 Acro-osteolysis syndrome, 565 Abruzzo-Erickson syndrome, 527, 538 Acro-reno-ocular syndrome, 527, Acanthosis, 607 539 Acanthosis nigricans, Crouzon Acyclovir, 504, 505, 518 syndrome-related, 169 Adam complex, 207–209 Physician-parent interactions, Adams-Oliver syndrome, 527, 540 69–70 Adduction, Möbius sequence-related Acetyl-coenzyme A deficiency, 364 impairment of, 197, 198 N-Acetylcystine, 321 Adenoma sebaceum, tuberous sclerosis N-Acetyl galactosamine-4-sulfatase complex-related, 304, 308–309 deficiency, 366 Adrenocorticotropic hormone, 433 N-Acetyl galactosamine-6-sulfatase Aging, premature. See also Progeria deficiency, 357, 365 ataxia-telangiectasia-related, 320 a-N-Acetyl glucosaminidase acetyl Aglossia-adactyly syndrome, 584–585 transferase deficiency, 357 Agnathia-holoprosencephaly, 54 a-N-Acetyl glucosaminidase -

BIRTH DEFECTS COMPENDIUM Second Edition BIRTH DEFECTS COMPENDIUM Second Edition
BIRTH DEFECTS COMPENDIUM Second Edition BIRTH DEFECTS COMPENDIUM Second Edition Editor Daniel Bergsma, MD, MPH Clinical Professor of Pediatrics Tufts University, School of Medicine Boston, Massachusetts * * * M Palgrave Macmillan ©The National Foundation 1973,1979 Softcover reprint of the hardcover 1st edition 1979 978-0-333-27876-5 All rights reserved. No part of this publication may be reproduced or transmitted, in any form or by any means, without permission. First published in the U.S.A. 1973, as Birth Defects Atlas and Compendium, by The Williams and Wilkins Company. Reprinted 1973,1974. Second Edition, published by Alan R. Liss, Inc., 1979. First published in the United Kingdom 1979 by THE MACMILLAN PRESS LTD London and Basingstoke Associated companies in Delhi Dublin Hong Kong Johannesburg Lagos Melbourne New York Singapore and Tokyo ISBN 978-1-349-05133-5 ISBN 978-1-349-05131-1 (eBook) DOI 10.1007/978-1-349-05131-1 Views expressed in articles published are the authors', and are not to be attributed to The National Foundation or its editors unless expressly so stated. To enhance medical communication in the birth defects field, The National Foundation has published the Birth Defects Atlas and Compendium, Syndrome ldentification, Original Article Series and developed a series of films and related brochures. Further information can be obtained from: The National Foundation- March of Dimes 1275 Mamaroneck Avenue White Plains, New York 10605 This book is sold subject to the standard conditions of the Net Book Agreement. DEDICATED To each dear little child who is in need of special help and care: to each eager parent who is desperately, hopefully seeking help: to each professional who brings understanding, knowledge and skillful care: to each generous friend who assists The National Foundation to help. -
ORD Resources Report
Resources and their URL's 12/1/2013 Resource Name: Resource URL: 1 in 9: The Long Island Breast Cancer Action Coalition http://www.1in9.org 11q Research and Resource Group http://www.11qusa.org 1p36 Deletion Support & Awareness http://www.1p36dsa.org 22q11 Ireland http://www.22q11ireland.org 22qcentral.org http://22qcentral.org 2q23.org http://2q23.org/ 4p- Support Group http://www.4p-supportgroup.org/ 4th Angel Mentoring Program http://www.4thangel.org 5p- Society http://www.fivepminus.org A Foundation Building Strength http://www.buildingstrength.org A National Support group for Arthrogryposis Multiplex http://www.avenuesforamc.com Congenita (AVENUES) A Place to Remember http://www.aplacetoremember.com/ Aarons Ohtahara http://www.ohtahara.org/ About Special Kids http://www.aboutspecialkids.org/ AboutFace International http://aboutface.ca/ AboutFace USA http://www.aboutfaceusa.org Accelerate Brain Cancer Cure http://www.abc2.org Accelerated Cure Project for Multiple Sclerosis http://www.acceleratedcure.org Accord Alliance http://www.accordalliance.org/ Achalasia 101 http://achalasia.us/ Acid Maltase Deficiency Association (AMDA) http://www.amda-pompe.org Acoustic Neuroma Association http://anausa.org/ Addison's Disease Self Help Group http://www.addisons.org.uk/ Adenoid Cystic Carcinoma Organization International http://www.accoi.org/ Adenoid Cystic Carcinoma Research Foundation http://www.accrf.org/ Advocacy for Neuroacanthocytosis Patients http://www.naadvocacy.org Advocacy for Patients with Chronic Illness, Inc. http://www.advocacyforpatients.org -

EUROCAT Syndrome Guide
JRC - Central Registry european surveillance of congenital anomalies EUROCAT Syndrome Guide Definition and Coding of Syndromes Version July 2017 Revised in 2016 by Ingeborg Barisic, approved by the Coding & Classification Committee in 2017: Ester Garne, Diana Wellesley, David Tucker, Jorieke Bergman and Ingeborg Barisic Revised 2008 by Ingeborg Barisic, Helen Dolk and Ester Garne and discussed and approved by the Coding & Classification Committee 2008: Elisa Calzolari, Diana Wellesley, David Tucker, Ingeborg Barisic, Ester Garne The list of syndromes contained in the previous EUROCAT “Guide to the Coding of Eponyms and Syndromes” (Josephine Weatherall, 1979) was revised by Ingeborg Barisic, Helen Dolk, Ester Garne, Claude Stoll and Diana Wellesley at a meeting in London in November 2003. Approved by the members EUROCAT Coding & Classification Committee 2004: Ingeborg Barisic, Elisa Calzolari, Ester Garne, Annukka Ritvanen, Claude Stoll, Diana Wellesley 1 TABLE OF CONTENTS Introduction and Definitions 6 Coding Notes and Explanation of Guide 10 List of conditions to be coded in the syndrome field 13 List of conditions which should not be coded as syndromes 14 Syndromes – monogenic or unknown etiology Aarskog syndrome 18 Acrocephalopolysyndactyly (all types) 19 Alagille syndrome 20 Alport syndrome 21 Angelman syndrome 22 Aniridia-Wilms tumor syndrome, WAGR 23 Apert syndrome 24 Bardet-Biedl syndrome 25 Beckwith-Wiedemann syndrome (EMG syndrome) 26 Blepharophimosis-ptosis syndrome 28 Branchiootorenal syndrome (Melnick-Fraser syndrome) 29 CHARGE -

Cockayne Syndrome
NLM Citation: Laugel V. Cockayne Syndrome. 2000 Dec 28 [Updated 2019 Aug 29]. In: Adam MP, Ardinger HH, Pagon RA, et al., editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993-2020. Bookshelf URL: https://www.ncbi.nlm.nih.gov/books/ Cockayne Syndrome Vincent Laugel, MD, PhD1 Created: December 28, 2000; Updated: August 29, 2019. Summary Clinical characteristics Cockayne syndrome (referred to as CS in this GeneReview) spans a continuous phenotypic spectrum that includes: • CS type I, the "classic" or "moderate" form; • CS type II, a more severe form with symptoms present at birth; this form overlaps with cerebrooculofacioskeletal (COFS) syndrome; • CS type III, a milder and later-onset form; • COFS syndrome, a fetal form of CS. CS type I is characterized by normal prenatal growth with the onset of growth and developmental abnormalities in the first two years. By the time the disease has become fully manifest, height, weight, and head circumference are far below the fifth percentile. Progressive impairment of vision, hearing, and central and peripheral nervous system function leads to severe disability; death typically occurs in the first or second decade. CS type II is characterized by growth failure at birth, with little or no postnatal neurologic development. Congenital cataracts or other structural anomalies of the eye may be present. Affected children have early postnatal contractures of the spine (kyphosis, scoliosis) and joints. Death usually occurs by age five years. CS type III is a phenotype in which major clinical features associated with CS only become apparent after age two years; growth and/or cognition exceeds the expectations for CS type I.