The Intergenic Spacer Region of the Rdna in Haplopappus Gracilis (Nutt.) Gray
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Notes on Variation and Geography in Rayjacksonia Phyllocephala (Asteraceae: Astereae)
Nesom, G.L., D.J. Rosen, and S.K. Lawrence. 2013. Notes on variation and geography in Rayjacksonia phyllocephala (Asteraceae: Astereae). Phytoneuron 2013-53: 1–15. Published 12 August 2013. ISSN 2153 733X NOTES ON VARIATION AND GEOGRAPHY IN RAYJACKSONIA PHYLLOCEPHALA (ASTERACEAE: ASTEREAE) GUY L. NESOM 2925 Hartwood Drive Fort Worth, Texas 76109 [email protected] DAVID J. ROSEN Department of Biology Lee College Baytown, Texas 77522-0818 [email protected] SHIRON K. LAWRENCE Department of Biology Lee College Baytown, Texas 77522-0818 ABSTRACT Inflorescences of Rayjacksonia phyllocephala in the disjunct Florida population system are characterized by heads on peduncles with leaves mostly reduced to linear bracts; heads in inflorescences of the Mexico-Texas-Louisiana system are immediately subtended by relatively unreduced leaves. The difference is consistent and justifies recognition of the Florida system as R. phyllocephala var. megacephala (Nash) D.B. Ward. Scattered waifs between the two systems are identified here as one or the other variety, directly implying their area of origin. In the eastern range of var. phyllocephala , at least from Brazoria County, Texas, eastward about 300 miles to central Louisiana, leaf margins vary from entire to deeply toothed-spinulose. In contrast, margins are invariably toothed-spinulose in var. megacephala as well as in the rest of southeastern Texas (from Brazoria County southwest) into Tamaulipas, Mexico. In some of the populations with variable leaf margins, 70-95% of the individuals have entire to mostly entire margins. KEY WORDS : Rayjacksonia , morphological variation, leaf margins, disjunction, waifs Rayjacksonia phyllocephala (DC.) Hartman & Lane (Gulf Coast camphor-daisy) is an abundant and conspicuous species of the shore vegetation around the Gulf of Mexico. -
1 C M 2 B H 3 B M 4 C M 5 B L 6 B M 7 B M 8
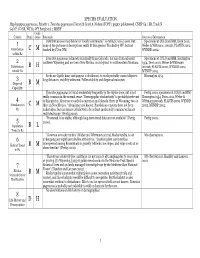
SPECIES EVALUATION Haplopappus pygmaeus, Priority 1. Tonestus pygmaeus (Torrey & Gray) A. Nelson (TOPY). pygmy goldenweed. CNHP G4 / SR, Track N G4 N?. CO SR, WY S1. WY Peripheral 2 MBNF Confi- Criteria Rank dence Rationale Sources of Information Distribution (see map below) is “nearly continuous,” so rating C was chosen, but Specimens at COLO and RM, Dorn 2001, 1 none of the pictures or descriptions really fit this species. Tracked by WY, but not Weber & Wittmann 2001ab, PLANTS 2002, Distribution C M tracked by CO or NM. WYNDD 2002. within R2 Tonestus pygmaeus is known principally from Colorado, but also from adjacent Specimens at COLO and RM, Harrington 2 southern Wyoming and northern New Mexico, and disjunct in southwestern Montana. 1954, Dorn 2001, Weber & Wittmann Distribution B H 2001ab, PLANTS 2002, WYNDD 2002, outside R2 MTNHP 2002. Seeds are lightly hairy and pappus is deciduous, so seeds probably cannot disperse Harrington 1954. 3 long distances; viability unknown. Pollen viability and dispersal unknown. Dispersal B M Capability Tonestus pygmaeus is found moderately frequently in the Alpine zone, but is not Fertig 2000, specimens at COLO and RM, really common in the normal sense. “Demographic stochasticity” is probably irrelevant Harrington 1954, Dorn 2001, Weber & 4 to this species. About 50 recorded occurrences in Colorado, three in Wyoming, two or Wittmann 2001ab, PLANTS 2002, WYNDD Abundance in C M three in New Mexico. “Abundance not known. Population censuses have not been 2002, MTNHP 2002. R2 undertaken, but the species is believed to be at least moderately common within its restricted range” (Fertig 2000). -

Reclassification of North American Haplopappus (Compositae: Astereae) Completed: Rayjacksonia Gen
AmericanJournal of Botany 83(3): 356-370. 1996. RECLASSIFICATION OF NORTH AMERICAN HAPLOPAPPUS (COMPOSITAE: ASTEREAE) COMPLETED: RAYJACKSONIA GEN. NOV.1 MEREDITH A. LANE2 AND RONALD L. HARTMAN R. L. McGregor Herbarium(University of Kansas NaturalHistory Museum Division of Botany) and Departmentof Botany,University of Kansas, Lawrence, Kansas 66047-3729; and Rocky MountainHerbarium, Department of Botany,University of Wyoming,Laramie, Wyoming82071-3165 Rayjacksonia R. L. Hartman& M. A. Lane, gen. nov. (Compositae: Astereae), is named to accommodate the "phyllo- cephalus complex," formerlyof Haplopappus Cass. sect. Blepharodon DC. The new combinationsare R. phyllocephalus (DC.) R. L. Hartman& M. A. Lane, R. annua (Rydb.) R. L. Hartman& M. A. Lane, and R. aurea (A. Gray) R. L. Hartman & M. A. Lane. This transfercompletes the reclassificationof the North American species of Haplopappus sensu Hall, leaving that genus exclusively South American.Rayjacksonia has a base chromosomenumber of x = 6. Furthermore,it shares abruptlyampliate disk corollas, deltatedisk style-branchappendages, and corolla epidermalcell type,among other features,with Grindelia, Isocoma, Olivaea, Prionopsis, Stephanodoria, and Xanthocephalum.Phylogenetic analyses of morphologicaland chloroplastDNA restrictionsite data, taken together,demonstrate that these genera are closely related but distinct. Key words: Astereae; Asteraceae; Compositae; Haplopappus; Rayjacksonia. During the past seven decades, taxonomic application lopappus sensu Hall (1928) are reclassifiedand are cur- -

Vascular Plants and a Brief History of the Kiowa and Rita Blanca National Grasslands
United States Department of Agriculture Vascular Plants and a Brief Forest Service Rocky Mountain History of the Kiowa and Rita Research Station General Technical Report Blanca National Grasslands RMRS-GTR-233 December 2009 Donald L. Hazlett, Michael H. Schiebout, and Paulette L. Ford Hazlett, Donald L.; Schiebout, Michael H.; and Ford, Paulette L. 2009. Vascular plants and a brief history of the Kiowa and Rita Blanca National Grasslands. Gen. Tech. Rep. RMRS- GTR-233. Fort Collins, CO: U.S. Department of Agriculture, Forest Service, Rocky Mountain Research Station. 44 p. Abstract Administered by the USDA Forest Service, the Kiowa and Rita Blanca National Grasslands occupy 230,000 acres of public land extending from northeastern New Mexico into the panhandles of Oklahoma and Texas. A mosaic of topographic features including canyons, plateaus, rolling grasslands and outcrops supports a diverse flora. Eight hundred twenty six (826) species of vascular plant species representing 81 plant families are known to occur on or near these public lands. This report includes a history of the area; ethnobotanical information; an introductory overview of the area including its climate, geology, vegetation, habitats, fauna, and ecological history; and a plant survey and information about the rare, poisonous, and exotic species from the area. A vascular plant checklist of 816 vascular plant taxa in the appendix includes scientific and common names, habitat types, and general distribution data for each species. This list is based on extensive plant collections and available herbarium collections. Authors Donald L. Hazlett is an ethnobotanist, Director of New World Plants and People consulting, and a research associate at the Denver Botanic Gardens, Denver, CO. -

Jessica's Aster
Jessica’s Aster (Aster jessicae) Population Monitoring: Third-year Results Idaho Conservation Data Center Idaho Department of Fish and Game PO Box 25 Juanita Lichthardt Boise, Idaho Karen Gray 83707 2005 Report prepared for the U.S. Fish and Wildlife Service ABSTRACT Jessica’s aster (Aster jessicae) is a tall, rhizomatous aster endemic to the Palouse region of southeastern Washington and adjacent Idaho. Its habitat has been severely reduced by conversion of this region to intensive agricultural uses. It is restricted almost entirely to private lands, and because of this, little was known of its distribution and abundance until a 1991 status survey was conducted. Five permanent monitoring plots were established in 2001, to track and compare populations occupying contrasting sites. Monitoring sites differ in amount of edge, degree of isolation from other populations, habitat extent, and amount of forest cover. This report summarizes three consecutive years of monitoring data. i TABLE OF CONTENTS ABSTRACT ......................................................................................................................................i TABLE OF CONTENTS ................................................................................................................ ii LIST OF TABLES ......................................................................................................................... iii LIST OF FIGURES....................................................................................................................... -

Baja California, Mexico, and a Vegetation Map of Colonet Mesa Alan B
Aliso: A Journal of Systematic and Evolutionary Botany Volume 29 | Issue 1 Article 4 2011 Plants of the Colonet Region, Baja California, Mexico, and a Vegetation Map of Colonet Mesa Alan B. Harper Terra Peninsular, Coronado, California Sula Vanderplank Rancho Santa Ana Botanic Garden, Claremont, California Mark Dodero Recon Environmental Inc., San Diego, California Sergio Mata Terra Peninsular, Coronado, California Jorge Ochoa Long Beach City College, Long Beach, California Follow this and additional works at: http://scholarship.claremont.edu/aliso Part of the Biodiversity Commons, Botany Commons, and the Ecology and Evolutionary Biology Commons Recommended Citation Harper, Alan B.; Vanderplank, Sula; Dodero, Mark; Mata, Sergio; and Ochoa, Jorge (2011) "Plants of the Colonet Region, Baja California, Mexico, and a Vegetation Map of Colonet Mesa," Aliso: A Journal of Systematic and Evolutionary Botany: Vol. 29: Iss. 1, Article 4. Available at: http://scholarship.claremont.edu/aliso/vol29/iss1/4 Aliso, 29(1), pp. 25–42 ’ 2011, Rancho Santa Ana Botanic Garden PLANTS OF THE COLONET REGION, BAJA CALIFORNIA, MEXICO, AND A VEGETATION MAPOF COLONET MESA ALAN B. HARPER,1 SULA VANDERPLANK,2 MARK DODERO,3 SERGIO MATA,1 AND JORGE OCHOA4 1Terra Peninsular, A.C., PMB 189003, Suite 88, Coronado, California 92178, USA ([email protected]); 2Rancho Santa Ana Botanic Garden, 1500 North College Avenue, Claremont, California 91711, USA; 3Recon Environmental Inc., 1927 Fifth Avenue, San Diego, California 92101, USA; 4Long Beach City College, 1305 East Pacific Coast Highway, Long Beach, California 90806, USA ABSTRACT The Colonet region is located at the southern end of the California Floristic Province, in an area known to have the highest plant diversity in Baja California. -

Vascular Flora of West Clear Creek Wilderness, Coconino and Yavapai
VASCULAR FLORA OF WEST CLEAR CREEK WILDERNESS, COCONINO AND YAVAPAI COUNTIES, ARIZONA By Wendy C. McBride A Thesis Submitted in Partial Fulfillment of the Requirements for the Degree of Master of Science in Biology Northern Arizona University May 2016 Approved: Tina J. Ayers, Ph.D., Chair Randall W. Scott, Ph.D. Liza M. Holeski, Ph.D. ABSTRACT VASCULAR FLORA OF WEST CLEAR CREEK WILDERNESS, COCONINO AND YAVAPAI COUNTIES, ARIZONA WENDY C. MCBRIDE West Clear Creek Wilderness bisects the Mogollon Rim in Arizona, and is nested between the Colorado Plateau and Basin and Range physiographic provinces. Between 2013 and 2016, a floristic inventory vouchered 542 taxa and reviewed 428 previous collections to produce a total plant inventory of 594 taxa from 93 families and 332 genera. The most species rich families Were Asteraceae, Poaceae, Fabaceae, Brassicaceae, Rosaceae, Plantaginaceae, Cyperaceae, and Polygonaceae. Carex, Erigeron, Bromus, Muhlenbergia, and Oenothera Were the most represented genera. Nonnative taxa accounted for seven percent of the total flora. Stachys albens was vouchered as a new state record for Arizona. New county records include Graptopetalum rusbyi (Coconino), Pseudognaphalium pringlei (Coconino), Phaseolus pedicellatus var. grayanus (Coconino), and Quercus rugosa (Coconino and Yavapai). This study quantified and contrasted native species diversity in canyon versus non- canyon floras across the Southwest. Analyses based on eighteen floras indicate that those centered about a major canyon feature shoW greater diversity than non-canyon floras. Regression models revealed that presence of a canyon Was a better predictor of similarity between floras than was the distance betWeen them. This study documents the remarkable diversity found Within canyon systems and the critical, yet varied, habitat they provide in the southwestern U.S. -

FLORATECH New Crop Report
FLORATECH New Crop Report presents: Machaeranthera gracilis Slender Goldenweed Taxonomy Machaeranthera gracilis (Nutt.) Shinners Translated: sword-like anthers; graceful. A.k.a.: Dieteria gracilis Haplopappus gracilis Haplopappus ravenii Sideranthus gracilis And newly named Xanthisma gracile WITHIN ASTERACEAE GEOGRAPHIC DISTRIBUTION Native to CA, NV, UT, AZ, CO, NM, TX, ME, NY according to USDA NRCS No reports of naturalizing Climactic Conditions5: Sandy or rocky places to 5000’ Habitats: creosote bush scrub, joshua tree woodland, east Mojave Desert Taxonomic Description7 Annual with erect stems, 3.25cm, leafy, branched at or above base, bristly throughout. Leaves: 1-3cm long, 3-7mm wide; lower oblanceolate, elliptic, or oblong in outline, 1-2 pinnately lobed; upper linear, reduced, lobes and teeth bristle-tipped. Inflorescence: heads radiate, solitary or in cymes; involucre 6-7mm, 7-12mm wide, hemispheric; phyllaries (bracts) in rows of 4-6, linear-lanceolate, bristle-tipped, hairy. Flowering time often from April-June. Ray flower: 16-18; ligules 7-12mm, yellow. Disk flower: 44-65; corollas 4.5-5.5mm, yellow. Taxonomic Description cont. Machaeranthera spp. tend to have several short rhizomes arising from the caudex (woody base of a perennial).1 Rhizomes may or may not function in vegetative reproduction.1 Biseasonal (germinates in winter, but flowers and sets seed in summer.2 Seed set greatly varied from season to season and shows significant responses to microhabitat differentiation.2 Use by indigenous people The Ramah Navaho used M. gracilis3: In a cold infusion used as lotion for pimples, boils and sores. In a cold, compound infusion of plant used as an eyewash. -

Jeffrey James Keeling Sul Ross State University Box C-64 Alpine, Texas 79832-0001, U.S.A
AN ANNOTATED VASCULAR FLORA AND FLORISTIC ANALYSIS OF THE SOUTHERN HALF OF THE NATURE CONSERVANCY DAVIS MOUNTAINS PRESERVE, JEFF DAVIS COUNTY, TEXAS, U.S.A. Jeffrey James Keeling Sul Ross State University Box C-64 Alpine, Texas 79832-0001, U.S.A. [email protected] ABSTRACT The Nature Conservancy Davis Mountains Preserve (DMP) is located 24.9 mi (40 km) northwest of Fort Davis, Texas, in the northeastern region of the Chihuahuan Desert and consists of some of the most complex topography of the Davis Mountains, including their summit, Mount Livermore, at 8378 ft (2554 m). The cool, temperate, “sky island” ecosystem caters to the requirements that are needed to accommo- date a wide range of unique diversity, endemism, and vegetation patterns, including desert grasslands and montane savannahs. The current study began in May of 2011 and aimed to catalogue the entire vascular flora of the 18,360 acres of Nature Conservancy property south of Highway 118 and directly surrounding Mount Livermore. Previous botanical investigations are presented, as well as biogeographic relation- ships of the flora. The numbers from herbaria searches and from the recent field collections combine to a total of 2,153 voucher specimens, representing 483 species and infraspecies, 288 genera, and 87 families. The best-represented families are Asteraceae (89 species, 18.4% of the total flora), Poaceae (76 species, 15.7% of the total flora), and Fabaceae (21 species, 4.3% of the total flora). The current study represents a 25.44% increase in vouchered specimens and a 9.7% increase in known species from the study area’s 18,360 acres and describes four en- demic and fourteen non-native species (four invasive) on the property. -

Asters of Yesteryear (Updated April 2018)
Asters of Yesteryear (Updated April 2018) About this Update: The document was originally posted in a shorter version, to accompany the brief article "Where Have all our Asters Gone?" in the Fall 2017 issue of Sego Lily. In that version it consisted simply of photos of a number of plants that had at some time been included in Aster but that no longer are, as per Flora of North America. In this version I have added names to the photos to indicate how they have changed since their original publication: Date and original name as published (Basionym) IF name used in Intermountain Flora (1994) UF name used in A Utah Flora (1983-2016) FNA name used in Flora of North America (2006) I have also added tables to show the renaming of two groups of species in the Astereae tribe as organized in Intermountain Flora. Color coding shows how splitting of the major genera largely follows fault lines already in place No color Renamed Bright Green Conserved Various Natural groupings $ Plant not in Utah It is noteworthy how few species retain the names used in 1994, but also how the renaming often follows patterns already observed. Asters of Yesteryear (Updated April 2018) Here are larger photos (16 inches wide or tall at normal screen resolution of 72 dpi) of the plants shown in Sego Lily of Fall 2017, arranged by date of original publication. None of them (except Aster amellus on this page) are now regarded as true asters – but they all were at one stage in their history. Now all are in different genera, most of them using names that were published over 100 years ago. -

Report on the Conservation Status of Haplopappus Radiatus, in Idaho
REPORT ON THE CONSERVATION STATUS OF HAPLOPAPPUS RADIATUS, IN IDAHO by Michael Mancuso and Robert K. Moseley Conservation Data Center Nongame and Endangered Wildlife Program June, 1993 Idaho Department of Fish and Game 600 South Walnut, P.O. Box 25 Boise, Idaho 83707 Jerry M. Conley, Director Status Survey Report prepared for Idaho Department of Parks and Recreation through Section 6 funding from U.S. Fish and Wildlife Service, Region 1 REPORT ON THE CONSERVATION STATUS OF i HAPLOPAPPUS RADIATUS, IN IDAHO Taxon Name: Haplopappus radiatus (Nutt.) Cronq. Common Name: Snake River goldenweed Family: Asteraceae (Compositae) States Where Taxon Occurs: U.S.A.; Idaho, Oregon Current Federal Status: Category 1 Candidate Recommended Federal Status: Threatened Authors of Report: Michael Mancuso and Robert K. Moseley Original Date of Report: June 18, 1993 Date of Most Recent Revision: N/A Individual to Whom Further Information and Comments Should be Sent: Robert K. Moseley Conservation Data Center Idaho Dept. Fish and Game P.O. Box 25 Boise, ID 83707 ii ABSTRACT Snake River goldenweed (Haplopappus radiatus) has been recognized as a possible conservation concern for at least twenty years. Until 1991, field inventories in Idaho were limited. In 1991, the Idaho Conservation Data Center completed a field investigation for Snake River goldenweed on the Payette National Forest. Through funding provided by the U.S. Fish and Wildlife Service, additional fieldwork was completed throughout the rest of the species' range in Idaho, during 1992. Survey work has also been done in Oregon. Besides field inventories, research biologists associated with the Conservation Biology Program, Oregon Department of Agriculture, have done cytological and pollination investigations, and presently are monitoring several populations in Oregon. -

2017 Colorado Rare Plant Symposia USFS Sensitive Species of Colorado Forest Service Status Species
2017 Colorado Rare Plant Symposia USFS Sensitive Species of Colorado Forest Service Status Species • National Forest Management Act (NFMA) – law that directs the management of NFS lands – NFMA is implemented in each National Forest’s or National Grassland’s Land Management Plan (LMP) • The way we do Land Management Plans has changed, status species lists will change accordingly – NFS units that have revised their LMP will have Species of Conservation Concern – Those that have not will retain Sensitive Species until they revise • Species protected under the Endangered Species Act are different Sensitive Species • What they are: Species with evidence of a current or predicted downward trend • Threshold for management: No decision can be made that will put a Sensitive species on a trend towards listing as Threatened or Endangered or would lead to the loss of viability on the planning unit (the area that the LMP covers) • Will continue to be updated until LMP is revised Species of Conservation Concern (SCC) • What they are: substantial concern about the species’ ability to persist over the long term in the plan area • Threshold for management: Effects of a proposed project can not result in ecological conditions that will not support SCC persistence within the plan area • Can be updated at any time after LMP revision Species of Conservation Concern • Management Focus: Creating or maintaining ecological conditions that promote recovery, conservation, and viability. These conditions are spelled out in the revised Land Management Plan. • Only NFS units that have revised their plans under the 2012 Rule will transition to SCC – Rio Grande NF – about halfway through process – Grand Mesa, Uncompahgre, and Gunnison NFs – just starting – The rest of the NFS units in Colorado are TBD and will keep Sensitive Species until they revise, sensitive list will continue to be updated Sensitive vs.