Protein Kinase C Epsilon Modulates Nicotine Consumption and Dopamine Reward Signals in the Nucleus Accumbens
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Gene Symbol Gene Description ACVR1B Activin a Receptor, Type IB
Table S1. Kinase clones included in human kinase cDNA library for yeast two-hybrid screening Gene Symbol Gene Description ACVR1B activin A receptor, type IB ADCK2 aarF domain containing kinase 2 ADCK4 aarF domain containing kinase 4 AGK multiple substrate lipid kinase;MULK AK1 adenylate kinase 1 AK3 adenylate kinase 3 like 1 AK3L1 adenylate kinase 3 ALDH18A1 aldehyde dehydrogenase 18 family, member A1;ALDH18A1 ALK anaplastic lymphoma kinase (Ki-1) ALPK1 alpha-kinase 1 ALPK2 alpha-kinase 2 AMHR2 anti-Mullerian hormone receptor, type II ARAF v-raf murine sarcoma 3611 viral oncogene homolog 1 ARSG arylsulfatase G;ARSG AURKB aurora kinase B AURKC aurora kinase C BCKDK branched chain alpha-ketoacid dehydrogenase kinase BMPR1A bone morphogenetic protein receptor, type IA BMPR2 bone morphogenetic protein receptor, type II (serine/threonine kinase) BRAF v-raf murine sarcoma viral oncogene homolog B1 BRD3 bromodomain containing 3 BRD4 bromodomain containing 4 BTK Bruton agammaglobulinemia tyrosine kinase BUB1 BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) BUB1B BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) C9orf98 chromosome 9 open reading frame 98;C9orf98 CABC1 chaperone, ABC1 activity of bc1 complex like (S. pombe) CALM1 calmodulin 1 (phosphorylase kinase, delta) CALM2 calmodulin 2 (phosphorylase kinase, delta) CALM3 calmodulin 3 (phosphorylase kinase, delta) CAMK1 calcium/calmodulin-dependent protein kinase I CAMK2A calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha CAMK2B calcium/calmodulin-dependent -

Endothelial Cell Injury in Atherosclerosis Is Regulated by Glycolysis (Review)
INTERNATIONAL JOURNAL OF MOleCular meDICine 47: 65-76, 2021 Mechanism overview and target mining of atherosclerosis: Endothelial cell injury in atherosclerosis is regulated by glycolysis (Review) RUIYING WANG1-5, MIN WANG1-5, JINGXUE YE1-5, GUIBO SUN1-5 and XIAOBO SUN1-5 1Institute of Medicinal Plant Development, Peking Union Medical College and Chinese Academy of Medical Sciences; 2Beijing Key Laboratory of Innovative Drug Discovery of Traditional Chinese Medicine (Natural Medicine) and Translational Medicine, Institute of Medicinal Plant Development, Peking Union Medical College and Chinese Academy of Medical Sciences; 3Key Laboratory of Bioactive Substances and Resources Utilization of Chinese Herbal Medicine, Ministry of Education, Institute of Medicinal Plant Development, Peking Union Medical College and Chinese Academy of Medical Sciences; 4Key Laboratory of Efficacy Evaluation of Chinese Medicine Against Glycolipid Metabolic Disorders, State Administration of Traditional Chinese Medicine, Institute of Medicinal Plant Development, Peking Union Medical College and Chinese Academy of Medical Sciences; 5Key Laboratory of New Drug Discovery Based on Classic Chinese Medicine Prescription, Chinese Academy of Medical Sciences, Beijing 100193, P.R. China Received August 14, 2020; Accepted November 5, 2020 DOI: 10.3892/ijmm.2020.4798 Abstract. Atherosclerosis (AS) is a chronic disease with a In conclusion, endothelial cell injury in AS may be alleviated by complex pathology that may lead to several cardiovascular and glycolysis and is a potential clinical treatment strategy for AS. cerebrovascular diseases; however, further research is neces- sary to fully elucidate its pathogenesis. The main risk factors for AS include lipid metabolism disorders, endothelial cell Contents injury, inflammation and immune dysfunction, among which vascular endothelial cell damage is considered as the main 1. -

Deep Multiomics Profiling of Brain Tumors Identifies Signaling Networks
ARTICLE https://doi.org/10.1038/s41467-019-11661-4 OPEN Deep multiomics profiling of brain tumors identifies signaling networks downstream of cancer driver genes Hong Wang 1,2,3, Alexander K. Diaz3,4, Timothy I. Shaw2,5, Yuxin Li1,2,4, Mingming Niu1,4, Ji-Hoon Cho2, Barbara S. Paugh4, Yang Zhang6, Jeffrey Sifford1,4, Bing Bai1,4,10, Zhiping Wu1,4, Haiyan Tan2, Suiping Zhou2, Laura D. Hover4, Heather S. Tillman 7, Abbas Shirinifard8, Suresh Thiagarajan9, Andras Sablauer 8, Vishwajeeth Pagala2, Anthony A. High2, Xusheng Wang 2, Chunliang Li 6, Suzanne J. Baker4 & Junmin Peng 1,2,4 1234567890():,; High throughput omics approaches provide an unprecedented opportunity for dissecting molecular mechanisms in cancer biology. Here we present deep profiling of whole proteome, phosphoproteome and transcriptome in two high-grade glioma (HGG) mouse models driven by mutated RTK oncogenes, PDGFRA and NTRK1, analyzing 13,860 proteins and 30,431 phosphosites by mass spectrometry. Systems biology approaches identify numerous master regulators, including 41 kinases and 23 transcription factors. Pathway activity computation and mouse survival indicate the NTRK1 mutation induces a higher activation of AKT down- stream targets including MYC and JUN, drives a positive feedback loop to up-regulate multiple other RTKs, and confers higher oncogenic potency than the PDGFRA mutation. A mini-gRNA library CRISPR-Cas9 validation screening shows 56% of tested master regulators are important for the viability of NTRK-driven HGG cells, including TFs (Myc and Jun) and metabolic kinases (AMPKa1 and AMPKa2), confirming the validity of the multiomics inte- grative approaches, and providing novel tumor vulnerabilities. -

The Human Gene Connectome As a Map of Short Cuts for Morbid Allele Discovery
The human gene connectome as a map of short cuts for morbid allele discovery Yuval Itana,1, Shen-Ying Zhanga,b, Guillaume Vogta,b, Avinash Abhyankara, Melina Hermana, Patrick Nitschkec, Dror Friedd, Lluis Quintana-Murcie, Laurent Abela,b, and Jean-Laurent Casanovaa,b,f aSt. Giles Laboratory of Human Genetics of Infectious Diseases, Rockefeller Branch, The Rockefeller University, New York, NY 10065; bLaboratory of Human Genetics of Infectious Diseases, Necker Branch, Paris Descartes University, Institut National de la Santé et de la Recherche Médicale U980, Necker Medical School, 75015 Paris, France; cPlateforme Bioinformatique, Université Paris Descartes, 75116 Paris, France; dDepartment of Computer Science, Ben-Gurion University of the Negev, Beer-Sheva 84105, Israel; eUnit of Human Evolutionary Genetics, Centre National de la Recherche Scientifique, Unité de Recherche Associée 3012, Institut Pasteur, F-75015 Paris, France; and fPediatric Immunology-Hematology Unit, Necker Hospital for Sick Children, 75015 Paris, France Edited* by Bruce Beutler, University of Texas Southwestern Medical Center, Dallas, TX, and approved February 15, 2013 (received for review October 19, 2012) High-throughput genomic data reveal thousands of gene variants to detect a single mutated gene, with the other polymorphic genes per patient, and it is often difficult to determine which of these being of less interest. This goes some way to explaining why, variants underlies disease in a given individual. However, at the despite the abundance of NGS data, the discovery of disease- population level, there may be some degree of phenotypic homo- causing alleles from such data remains somewhat limited. geneity, with alterations of specific physiological pathways under- We developed the human gene connectome (HGC) to over- come this problem. -

The Akt-Mtor Pathway Drives Myelin Sheath Growth by Regulating Cap-Dependent Translation
bioRxiv preprint doi: https://doi.org/10.1101/2021.04.12.439555; this version posted April 12, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC 4.0 International license. Title Page Manuscript Title: The Akt-mTOR pathway drives myelin sheath growth by regulating cap-dependent translation Abbreviated Title: mTOR-mediated translation drives myelination Authors: Karlie N. Fedder-Semmes1,2, Bruce Appel1,3 Affiliations: 1Department of Pediatrics, Section of Developmental Biology 2Neuroscience Graduate Program 3Children’s Hospital Colorado University of Colorado Anschutz Medical Campus Aurora, Colorado, United States of America, 80045 Corresponding author: Bruce Appel, [email protected] Number of pages: 77, including references and figures Number of figures: 8 Acknowledgements: We thank the Appel lab for helpful discussions, Drs. Caleb Doll and Alexandria Hughes for constructive comments on the manuscript, Dr. Katie Yergert for experimental guidance, and Rebecca O’Rourke for help with bioinformatics analysis. This work was supported by US National Institutes of Health (NIH) grant R01 NS095670 and a gift from the Gates Frontiers Fund to B.A. K.N.F-S. was supported by NIH F31 NS115261 and NIH T32 NS099042. The University of Colorado Anschutz Medical Campus Zebrafish Core Facility was supported by NIH grant P30 NS048154. Conflict of Interest: The authors declare no competing financial interests. bioRxiv preprint doi: https://doi.org/10.1101/2021.04.12.439555; this version posted April 12, 2021. -

Clinical, Molecular, and Immune Analysis of Dabrafenib-Trametinib
Supplementary Online Content Chen G, McQuade JL, Panka DJ, et al. Clinical, molecular and immune analysis of dabrafenib-trametinib combination treatment for metastatic melanoma that progressed during BRAF inhibitor monotherapy: a phase 2 clinical trial. JAMA Oncology. Published online April 28, 2016. doi:10.1001/jamaoncol.2016.0509. eMethods. eReferences. eTable 1. Clinical efficacy eTable 2. Adverse events eTable 3. Correlation of baseline patient characteristics with treatment outcomes eTable 4. Patient responses and baseline IHC results eFigure 1. Kaplan-Meier analysis of overall survival eFigure 2. Correlation between IHC and RNAseq results eFigure 3. pPRAS40 expression and PFS eFigure 4. Baseline and treatment-induced changes in immune infiltrates eFigure 5. PD-L1 expression eTable 5. Nonsynonymous mutations detected by WES in baseline tumors This supplementary material has been provided by the authors to give readers additional information about their work. © 2016 American Medical Association. All rights reserved. Downloaded From: https://jamanetwork.com/ on 09/30/2021 eMethods Whole exome sequencing Whole exome capture libraries for both tumor and normal samples were constructed using 100ng genomic DNA input and following the protocol as described by Fisher et al.,3 with the following adapter modification: Illumina paired end adapters were replaced with palindromic forked adapters with unique 8 base index sequences embedded within the adapter. In-solution hybrid selection was performed using the Illumina Rapid Capture Exome enrichment kit with 38Mb target territory (29Mb baited). The targeted region includes 98.3% of the intervals in the Refseq exome database. Dual-indexed libraries were pooled into groups of up to 96 samples prior to hybridization. -

Targeting the Hsp90-Cdc37-Client Protein Interaction to Disrupt Hsp90 Chaperone Machinery Ting Li1, Hu-Lin Jiang2, Yun-Guang Tong3,4 and Jin-Jian Lu1*
Li et al. Journal of Hematology & Oncology (2018) 11:59 https://doi.org/10.1186/s13045-018-0602-8 REVIEW Open Access Targeting the Hsp90-Cdc37-client protein interaction to disrupt Hsp90 chaperone machinery Ting Li1, Hu-Lin Jiang2, Yun-Guang Tong3,4 and Jin-Jian Lu1* Abstract Heat shock protein 90 (Hsp90) is a critical molecular chaperone protein that regulates the folding, maturation, and stability of a wide variety of proteins. In recent years, the development of Hsp90-directed inhibitors has grown rapidly, and many of these inhibitors have entered clinical trials. In parallel, the functional dissection of the Hsp90 chaperone machinery has highlighted the activity disruption of Hsp90 co-chaperone as a potential target. With the roles of Hsp90 co-chaperones being elucidated, cell division cycle 37 (Cdc37), a ubiquitous co-chaperone of Hsp90 that directs the selective client proteins into the Hsp90 chaperone cycle, shows great promise. Moreover, the Hsp90-Cdc37-client interaction contributes to the regulation of cellular response and cellular growth and is more essential to tumor tissues than normal tissues. Herein, we discuss the current understanding of the clients of Hsp90-Cdc37, the interaction of Hsp90-Cdc37-client protein, and the therapeutic possibilities of targeting Hsp90-Cdc37-client protein interaction as a strategy to inhibit Hsp90 chaperone machinery to present new insights on alternative ways of inhibiting Hsp90 chaperone machinery. Keywords: Hsp90 chaperone machinery, Cdc37, Kinase client, Protein interaction Background chloroplast HSP90C, mitochondrial TNFR-associated protein, Heat shock protein 90 (Hsp90) is a critically conserved and bacterial high-temperature protein G [2, 8]. In this protein and one of the major molecular chaperones within review,weusethetermHsp90torefertotheseHsp90 eukaryotic cells [1]. -

Presentation
Understanding Signaling Pathways by Modifying Sensitivity to PLX4720 in B-RAFV600E Melanoma Muska Hassan NCI-ICBP Summer Fellow Broad Institute of MIT and Harvard: Cancer Program Mentor: Cory Johannessen, Ph.D. PIs: Todd Golub and Levi Garraway What Is Melanoma? • Leading cause of skin cancer death • Originates in melanocytes • Melanocytes produce skin/hair pigmentation Anatomy of the Skin. 2010. About Melanoma. 18 July. 2011. <www.cancerfacts.com/GeneralContent/Melanoma/gen_overview.asp?CB=8> BRAFV600E Mutation • Mitogen-activated protein kinase (MAPK) is activated through phosphorylation of MEK ERKcell proliferation/ CRAF BRAF growth • B-RAF gene: proto-oncogene MEK • Gets mutated at codon 600 • 50-70% of metastatic melanomas harbor this mutation ERK • Mutant B-RAF constitutively activates the MAPK pathway • Inhibiting B-RAF with PLX4720, the MAPK pathway is turned off and tumors regress • Despite initially regressing, all tumors eventually developed drug resistance A Screen for Kinases that Bypass B-RAF Inhibition >2 st.dev. from average Average (all ORFs) CM Johannessen et al. Nature 000, 1-5 (2010) doi:10.1038/nature09627 How Do Candidate Genes Induce Resistance? Prioritization Screen (2 cell lines, 8-point GI50) Rank Gene COT 1 COT Resistance via MAPK CRAF BRAF 2 C-RAF pathway activation +P 3 CRKL 4 FGR MEK 5 PRKCE 6 PRKCH ? ERK 7 ERBB2 8 AXL 9 PAK3 Cell Lines: SKMEL28 and A375 (B-RAF Mutant) Hypothesis Hypothesis: Based on the results obtained from the screen, we hypothesized that PRKC ε, η, and θ reactivate the MAPK signaling pathway OR an alternative signaling pathway independent of the MAPK pathway. (2009), 36th Annual Meeting of the Arbeitsgemeinschaft Dermatologische Forschung (ADF). -
![(PRKCE) Mouse Monoclonal Antibody [Clone ID: OTI4G8] Product Data](https://docslib.b-cdn.net/cover/4674/prkce-mouse-monoclonal-antibody-clone-id-oti4g8-product-data-3294674.webp)
(PRKCE) Mouse Monoclonal Antibody [Clone ID: OTI4G8] Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for TA502415 PKC epsilon (PRKCE) Mouse Monoclonal Antibody [Clone ID: OTI4G8] Product data: Product Type: Primary Antibodies Clone Name: OTI4G8 Applications: FC, IF, IHC, WB Recommended Dilution: WB 1:1000~2000, IHC 1:150, IF 1:100, FLOW 1:100 Reactivity: Human, Monkey, Mouse, Rat Host: Mouse Isotype: IgG2b Clonality: Monoclonal Immunogen: Full length human recombinant protein of human PRKCE (NP_005391) produced in HEK293T cell. Formulation: PBS (PH 7.3) containing 1% BSA, 50% glycerol and 0.02% sodium azide. Concentration: 0.5 mg/ml Purification: Purified from mouse ascites fluids or tissue culture supernatant by affinity chromatography (protein A/G) Conjugation: Unconjugated Storage: Store at -20°C as received. Stability: Stable for 12 months from date of receipt. Predicted Protein Size: 83.5 kDa Gene Name: protein kinase C epsilon Database Link: NP_005391 Entrez Gene 18754 MouseEntrez Gene 29340 RatEntrez Gene 714533 MonkeyEntrez Gene 5581 Human Q02156 This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 6 PKC epsilon (PRKCE) Mouse Monoclonal Antibody [Clone ID: OTI4G8] – TA502415 Background: Protein kinase C (PKC) is a family of serine- and threonine-specific protein kinases that can be activated by calcium and the second messenger diacylglycerol. PKC family members phosphorylate a wide variety of protein targets and are known to be involved in diverse cellular signaling pathways. -

Variants in PRKCE and KLC1, Potential Negative Regulators of Type I Psoriasis
Variants in PRKCE and KLC1, Potential Negative Regulators of Type I Psoriasis Jianxiao Xing Taiyuan central hospital of shanxi medical university Ying Wang Taiyuan central hospital of shanxi medical university Xincheng Zhao Taiyuan central hospital of shanxi medical university Junqin Li Taiyuan central hospital of shanxi medical university Ruixia Hou Taiyuan central hospital of shanxi medical university Xuping Niu Taiyuan central university of shanxi medical university Guohua Yin Taiyuan central hospital of shanxi medical university Xinhua Li Taiyuan central hospital of shanxi medical university kaiming zhang ( [email protected] ) Taiyuan central hospital of shanxi medical university https://orcid.org/0000-0001-9525-9944 Research article Keywords: psoriasis, gene variants, PRKCE, KLC1 Posted Date: January 19th, 2021 DOI: https://doi.org/10.21203/rs.3.rs-147917/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Page 1/10 Abstract Background: Psoriasis is a multifactorial disease with a complex genetic predisposition. The pathophysiology of psoriasis is associated with genetic variants, especially in negative regulatory genes. To better characterize gene variants in psoriasis and identify the relationship between clinical characteristics and variant genes in its pathogenesis. DNA was extracted from 282 type psoriasis patients and puried, and 13 variable genes were amplied and sequenced using the Sanger method. Results: Among the 13 investigated genes, the variants frequencies of protein kinase C epsilon (PRKCE) (c.240T>C, 35.9% vs 47.7%, P< .05) and kinesin light chain 1 (KLC1) (c.216A>G, 2.9% vs 98.1%, P< .01) were signicantly lower in patients than in normal Asian individuals. -

Supplementary Material Identification of Motility-Associated Progesterone
10.1071/RD15492_AC © CSIRO 2017 Supplementary Material: Reproduction, Fertility and Development, 2017, 29(6), 1115–1129. Supplementary Material Identification of motility-associated progesterone-responsive differentially phosphorylated proteins V. Sagare-PatilA and D. ModiA,B AMolecular and Cellular Biology Laboratory, National Institute for Research in Reproductive Health, Indian Council of Medical Research, JM Street, Parel, Mumbai 400012, India. BCorresponding author. Email: [email protected]; [email protected] Supplementary Method Method for measuring total tyrosine kinase activity Total tyrosine kinase activity was assist using tyrosine kinase ELISA kit Capacitated and progesterone treated spermatozoa (10×106 /ml) were solubilized in lysis buffer (50 mM HEPES buffer, pH 7.4, containing 0.1 % TRITON X-100, 10% glycerol, 1 mM dithiothreitol , protease inhibitor and cocktail phosphatase inhibitor cocktail) provided in the kit, sonicated and centrifuged and protein content was estimated. Kinase activity was determined in the supernatants using a synthetic peptide polyGlu-Tyr (PGT) as substrate provided in the kit. Standard curves were run along with test samples in each experiment. The assay was done for three different pools of samples per concentration per time point. PTK substrate was reconstituted with PBS and incubated in multiwell plate overnight. After washing the wells were dried at 370C. For the PTK assay standard and test samples were incubated Tyrosine Kinase 0.3 mM ATP at room temperature for 30 minutes. After repeated washing with wash buffer the antibody conjugate (1:100 diluted) was added in each well and incubated for 30 minutes at room temperature. After extensive washing, freshly prepared peroxidase substrate solution was added to each well and incubate for exactly 7 minutes, in the dark. -
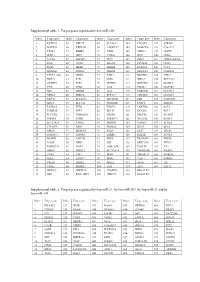
Target Genes Regulated by Hsa-Mir-21, by Hsa-Mir-203, by Hsa-Mir-21 and by Hsa-Mir-143
Supplemental table 1: Target genes regulated by hsa-miR-205 Index Target gene Index Target gene Index Target gene Index Target gene Index Target gene 1 KCTD20 35 UBE2Z 69 SLC38A1 103 LPCAT1 137 STK38L 2 MAPK14 36 YWHAH 70 ANGPTL7 104 MARCKS 138 C1orf123 3 TXNL1 37 RBBP4 71 CTGF 105 MED13 139 GUCD1 4 SPDL1 38 LRP1 72 CYR61 106 IPO7 140 CDK6 5 TCF20 39 IMPAD1 73 TP73 107 PHC2 141 CDKN2AIPNL 6 RAN 40 GNAS 74 EGLN2 108 PICALM 142 CLIP1 7 RGS6 41 MED1 75 ERBB2 109 PLAGL2 143 CUL5 8 HOXA11 42 INPPL1 76 PRRG4 110 NDUFA4 144 C6orf201 9 PAPPA-AS1 43 DDX5 77 F2RL2 111 NDUFB2 145 VTI1A 10 PRR15 44 E2F1 78 GOT1 112 NIPA2 146 SLC5A12 11 ACTRT3 45 E2F5 79 NUFIP2 113 NOTCH2 147 MAML2 12 YES1 46 ZEB2 80 IL24 114 PANK1 148 MAP3K9 13 SRC 47 ERBB3 81 IL32 115 PARD6B 149 NUDT21 14 NPRL3 48 PRKCE 82 RNF217 116 TMEM66 150 DNAJA1 15 NFAT5 49 SLC41A1 83 ZNF585B 117 EZR 151 CCDC108 16 XPOT 50 SLC7A2 84 SIGMAR1 118 ENPP4 152 SHISA6 17 KCTD16 51 ZEB1 85 VEGFA 119 LRRTM4 153 ACP1 18 TMSB4X 52 PHF8 86 BCL9L 120 KCNJ10 154 BCL2 19 PLCXD2 53 TMEM201 87 CREB1 121 PHLPP2 155 NCAPG 20 TNFSF8 54 PTPRJ 88 SERINC3 122 YEATS2 156 KLHL5 21 SLC25A25 55 ETNK1 89 HMGB3 123 VAMP1 157 ACSL4 22 C11orf74 56 XPR1 90 SRD5A1 124 RTN3 158 BCL6 23 GM2A 57 MRPL44 91 PTEN 125 RFX7 159 ITGA5 24 SMNDC1 58 TM9SF2 92 ESRRG 126 RAP2B 160 ACSL1 25 BAMBI 59 PAIP2B 93 PRLR 127 TRAF3IP1 161 EID2B 26 LCOR 60 NEK9 94 ICK 128 SERTAD2 162 TEX35 27 TMEM239 61 NOX5 95 LOH12CR1 129 TOLLIP 163 YY1 28 AMOT 62 DMXL2 96 SLC39A14 130 TMEM55B 164 SMAD1 29 CDK1 63 ETF1 97 BDP1 131 TMEM123 165 SMAD4 30 SQLE 64