Primary Cilia, Ciliogenesis and the Actin Cytoskeleton: a Little Less Resorption, a Little More Actin Please
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Download (PDF)
ANALYTICAL SCIENCES NOVEMBER 2020, VOL. 36 1 2020 © The Japan Society for Analytical Chemistry Supporting Information Fig. S1 Detailed MS/MS data of myoglobin. 17 2 ANALYTICAL SCIENCES NOVEMBER 2020, VOL. 36 Table S1 : The protein names (antigens) identified by pH 2.0 solution in the eluted-fraction. These proteins were identified one or more out of six analyses. Accession Description P08908 5-hydroxytryptamine receptor 1A OS=Homo sapiens GN=HTR1A PE=1 SV=3 - [5HT1A_HUMAN] Q9NRR6 72 kDa inositol polyphosphate 5-phosphatase OS=Homo sapiens GN=INPP5E PE=1 SV=2 - [INP5E_HUMAN] P82987 ADAMTS-like protein 3 OS=Homo sapiens GN=ADAMTSL3 PE=2 SV=4 - [ATL3_HUMAN] Q9Y6K8 Adenylate kinase isoenzyme 5 OS=Homo sapiens GN=AK5 PE=1 SV=2 - [KAD5_HUMAN] P02763 Alpha-1-acid glycoprotein 1 OS=Homo sapiens GN=ORM1 PE=1 SV=1 - [A1AG1_HUMAN] P19652 Alpha-1-acid glycoprotein 2 OS=Homo sapiens GN=ORM2 PE=1 SV=2 - [A1AG2_HUMAN] P01011 Alpha-1-antichymotrypsin OS=Homo sapiens GN=SERPINA3 PE=1 SV=2 - [AACT_HUMAN] P01009 Alpha-1-antitrypsin OS=Homo sapiens GN=SERPINA1 PE=1 SV=3 - [A1AT_HUMAN] P04217 Alpha-1B-glycoprotein OS=Homo sapiens GN=A1BG PE=1 SV=4 - [A1BG_HUMAN] P08697 Alpha-2-antiplasmin OS=Homo sapiens GN=SERPINF2 PE=1 SV=3 - [A2AP_HUMAN] P02765 Alpha-2-HS-glycoprotein OS=Homo sapiens GN=AHSG PE=1 SV=1 - [FETUA_HUMAN] P01023 Alpha-2-macroglobulin OS=Homo sapiens GN=A2M PE=1 SV=3 - [A2MG_HUMAN] P01019 Angiotensinogen OS=Homo sapiens GN=AGT PE=1 SV=1 - [ANGT_HUMAN] Q9NQ90 Anoctamin-2 OS=Homo sapiens GN=ANO2 PE=1 SV=2 - [ANO2_HUMAN] P01008 Antithrombin-III -

Synergistic Genetic Interactions Between Pkhd1 and Pkd1 Result in an ARPKD-Like Phenotype in Murine Models
BASIC RESEARCH www.jasn.org Synergistic Genetic Interactions between Pkhd1 and Pkd1 Result in an ARPKD-Like Phenotype in Murine Models Rory J. Olson,1 Katharina Hopp ,2 Harrison Wells,3 Jessica M. Smith,3 Jessica Furtado,1,4 Megan M. Constans,3 Diana L. Escobar,3 Aron M. Geurts,5 Vicente E. Torres,3 and Peter C. Harris 1,3 Due to the number of contributing authors, the affiliations are listed at the end of this article. ABSTRACT Background Autosomal recessive polycystic kidney disease (ARPKD) and autosomal dominant polycystic kidney disease (ADPKD) are genetically distinct, with ADPKD usually caused by the genes PKD1 or PKD2 (encoding polycystin-1 and polycystin-2, respectively) and ARPKD caused by PKHD1 (encoding fibrocys- tin/polyductin [FPC]). Primary cilia have been considered central to PKD pathogenesis due to protein localization and common cystic phenotypes in syndromic ciliopathies, but their relevance is questioned in the simple PKDs. ARPKD’s mild phenotype in murine models versus in humans has hampered investi- gating its pathogenesis. Methods To study the interaction between Pkhd1 and Pkd1, including dosage effects on the phenotype, we generated digenic mouse and rat models and characterized and compared digenic, monogenic, and wild-type phenotypes. Results The genetic interaction was synergistic in both species, with digenic animals exhibiting pheno- types of rapidly progressive PKD and early lethality resembling classic ARPKD. Genetic interaction be- tween Pkhd1 and Pkd1 depended on dosage in the digenic murine models, with no significant enhancement of the monogenic phenotype until a threshold of reduced expression at the second locus was breached. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

SUPPLEMENTARY MATERIAL Human Engineered Skeletal Muscle
SUPPLEMENTARY MATERIAL Human engineered skeletal muscle of hypaxial origin from pluripotent stem cells with advanced function and regenerative capacity Mina Shahriyari1,2, Md Rezaul Islam3, M. Sadman Sakib3, Anastasia Rika1,2, Dennis Krüger3, Lalit Kaurani3, Harithaa Anandakumar1,2, Orr Shomroni4, Matthias Schmidt5, Jana Zschüntzsch5, Jens Schmidt5, Gabriela Salinas-Riester4, Andreas Unger6, Wolfgang A. Linke6, André Fischer3,7, Wolfram-Hubertus Zimmermann1,2,3,7,8*, Malte Tiburcy1,2* 1 Institute of Pharmacology and Toxicology, University Medical Center Göttingen, Göttingen, Germany. 2 DZHK (German Center for Cardiovascular Research), partner site Göttingen. 3 Department for Epigenetics and Systems Medicine in Neurodegenerative Diseases, German Center for Neurodegenerative Diseases (DZNE) Göttingen, Göttingen, Germany 4 NGS Integrative Genomics Core Unit, Institute of Human Genetics, University Medical Center Göttingen, Göttingen, Germany 5 Department of Neurology, University Medical Center Göttingen, Göttingen, Germany 6 Institute of Physiology II, University of Münster, D-48149 Münster, Germany 7 Cluster of Excellence "Multiscale Bioimaging: from Molecular Machines to Networks of Excitable Cells" (MBExC), University of Göttingen, Germany; 8 Fraunhofer Institute for Translational Medicine and Pharmacology (ITMP), Göttingen, Germany Supplementary table 1 related to Figure 2: Biological process annotation of coexpression clusters. Cluster Biological process annotated Enriched GO terms identifier Black Migrating limb progenitors Muscle -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Effect of Fucoxanthinol on Pancreatic
CANCER GENOMICS & PROTEOMICS 18 : 407-423 (2021) doi:10.21873/cgp.20268 Effect of Fucoxanthinol on Pancreatic Ductal Adenocarcinoma Cells from an N-Nitrosobis(2-oxopropyl)amine-initiated Syrian Golden Hamster Pancreatic Carcinogenesis Model MASARU TERASAKI 1,2 , YUSAKU NISHIZAKA 1, WATARU MURASE 1, ATSUHITO KUBOTA 1, HIROYUKI KOJIMA 1,2 , MARESHIGE KOJOMA 1, TAKUJI TANAKA 3, HAYATO MAEDA 4, KAZUO MIYASHITA 5, MICHIHIRO MUTOH 6 and MAMI TAKAHASHI 7 1School of Pharmaceutical Sciences and 2Advanced Research Promotion Center, Health Sciences University of Hokkaido, Hokkaido, Japan; 3Department of Diagnostic Pathology and Research Center of Diagnostic Pathology, Gifu Municipal Hospital, Gifu, Japan; 4Faculty of Agriculture and Life Science, Hirosaki University, Aomori, Japan; 5Center for Industry-University Collaboration, Obihiro University of Agriculture and Veterinary Medicine, Obihiro, Japan; 6Department of Molecular-Targeting Prevention, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan; 7Central Animal Division, National Cancer Center, Tokyo, Japan Abstract. Background/Aim: Fucoxanthinol (FxOH) is a as a cancer chemopreventive agent in a hamster pancreatic marine carotenoid metabolite with potent anti-cancer activity. carcinogenesis model. However, little is known about the efficacy of FxOH in pancreatic cancer. In the present study, we investigated the Pancreatic cancer is one of the most lethal cancers worldwide inhibitory effect of FxOH on six types of cells cloned from N- because it is treatment resistant and has aggressive potential nitrosobis(2-oxopropyl)amine (BOP)-induced hamster for metastasis/invasion with resultant poor prognosis. From pancreatic cancer (HaPC) cells. Materials and Methods: the GLOBOCAN 2018 estimates, 432,242 pancreatic cancer FxOH action and its molecular mechanisms were investigated deaths occur per year (4.5% of total) (1), and the 5-year in HaPC cells using flow-cytometry, comprehensive gene survival rate remains poor at 10% (2 ). -

Post-Transcriptionally Impaired De Novo Mutations Contribute to The
bioRxiv preprint doi: https://doi.org/10.1101/175844; this version posted November 26, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Post-transcriptionally impaired de novo mutations 2 contribute to the genetic etiology of four neuropsychiatric 3 disorders 4 5 Fengbiao Mao1,2¶, Lu Wang3¶, Xiaolu Zhao2, Zhongshan Li4, Luoyuan Xiao5, 6 Rajesh C. Rao2, Jinchen Li4, Huajing Teng1*, Xin He6*, and Zhong Sheng Sun1,4* 7 8 1 Beijing Institutes of Life Science, Chinese Academy of Sciences, Beijing 100101, 9 China. 10 2 Department of Pathology, University of Michigan, Ann Arbor, MI 48109, USA. 11 3 Institute of Life Science, Southeast University, Nanjing 210096, China. 12 4 Institute of Genomic Medicine, Wenzhou Medical University, Wenzhou 325027, 13 China 14 5 Department of Computer Science and Technology, Tsinghua University, Beijing 15 100084, China. 16 6 Department of Human Genetics, University of Chicago, Chicago, IL, USA. 17 18 ¶These authors contributed equally to this work 19 * Corresponding authors 20 E-mail: 21 [email protected] (Z.S.S.) 22 [email protected] (X.H.) 23 [email protected] (H.T.) 24 25 1 bioRxiv preprint doi: https://doi.org/10.1101/175844; this version posted November 26, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

The Genetic Program of Pancreatic Beta-Cell Replication in Vivo
Page 1 of 65 Diabetes The genetic program of pancreatic beta-cell replication in vivo Agnes Klochendler1, Inbal Caspi2, Noa Corem1, Maya Moran3, Oriel Friedlich1, Sharona Elgavish4, Yuval Nevo4, Aharon Helman1, Benjamin Glaser5, Amir Eden3, Shalev Itzkovitz2, Yuval Dor1,* 1Department of Developmental Biology and Cancer Research, The Institute for Medical Research Israel-Canada, The Hebrew University-Hadassah Medical School, Jerusalem 91120, Israel 2Department of Molecular Cell Biology, Weizmann Institute of Science, Rehovot, Israel. 3Department of Cell and Developmental Biology, The Silberman Institute of Life Sciences, The Hebrew University of Jerusalem, Jerusalem 91904, Israel 4Info-CORE, Bioinformatics Unit of the I-CORE Computation Center, The Hebrew University and Hadassah, The Institute for Medical Research Israel- Canada, The Hebrew University-Hadassah Medical School, Jerusalem 91120, Israel 5Endocrinology and Metabolism Service, Department of Internal Medicine, Hadassah-Hebrew University Medical Center, Jerusalem 91120, Israel *Correspondence: [email protected] Running title: The genetic program of pancreatic β-cell replication 1 Diabetes Publish Ahead of Print, published online March 18, 2016 Diabetes Page 2 of 65 Abstract The molecular program underlying infrequent replication of pancreatic beta- cells remains largely inaccessible. Using transgenic mice expressing GFP in cycling cells we sorted live, replicating beta-cells and determined their transcriptome. Replicating beta-cells upregulate hundreds of proliferation- related genes, along with many novel putative cell cycle components. Strikingly, genes involved in beta-cell functions, namely glucose sensing and insulin secretion were repressed. Further studies using single molecule RNA in situ hybridization revealed that in fact, replicating beta-cells double the amount of RNA for most genes, but this upregulation excludes genes involved in beta-cell function. -

Focus on the Axon: from Neuronal Development to Dynamic Synapses ISBN: 978-90-393-6856-5
Focus on the axon: from neuronal development to dynamic synapses ISBN: 978-90-393-6856-5 The studies described in this thesis were performed at the division of Cell Biology at the Faculty of Science of the Utrecht University in Utrecht, the Netherlands. This thesis was partially supported by the People Programme (Marie Curie Actions; MC- ITN) of the European Union’s Seventh Framework Programme FP7/2007-2013/ under REA grant agreement 289581. The printing of the thesis was kindly supported by Vereniging van Huntington and Alzheimer Nederland. Cover: Neuron Tangram Cover design by Cátia P. Frias and Reinier Damman. Layout by Cátia P. Frias. Printed by Gildeprint. Copyright © Cátia Sofia Pereira Frias. All rights reserved. Focus on the axon: from neuronal development to dynamic synapses Het axon centraal: van ontwikkelende hersencellen tot dynamische synapsen (met een samenvatting in het Nederlands) Foco no axónio: do desenvolvimento neuronal às sinapses dinâmicas (com um sumário em Português) Proefschrift ter verkrijging van de graad van doctor aan de Universiteit Utrecht op gezag van de rector magnificus, prof.dr. G.J. van der Zwaan, ingevolge het besluit van het college voor promoties in het openbaar te verdedigen op woensdag 8 november 2017 des middags te 2.30 uur door Cátia Sofia Pereira Frias geboren op 15 januari 1989 te Funchal, Portugal Promotor: Prof. dr. C. C. Hoogenraad Copromotor: Dr. C. J. Wierenga Para os meus pais TABLE OF CONTENTS Chapter 1 General Introduction 9 Chapter 2 Activity-dependent adaptations in inhibitory axons -

Downloaded Per Proteome Cohort Via the Web- Site Links of Table 1, Also Providing Information on the Deposited Spectral Datasets
www.nature.com/scientificreports OPEN Assessment of a complete and classifed platelet proteome from genome‑wide transcripts of human platelets and megakaryocytes covering platelet functions Jingnan Huang1,2*, Frauke Swieringa1,2,9, Fiorella A. Solari2,9, Isabella Provenzale1, Luigi Grassi3, Ilaria De Simone1, Constance C. F. M. J. Baaten1,4, Rachel Cavill5, Albert Sickmann2,6,7,9, Mattia Frontini3,8,9 & Johan W. M. Heemskerk1,9* Novel platelet and megakaryocyte transcriptome analysis allows prediction of the full or theoretical proteome of a representative human platelet. Here, we integrated the established platelet proteomes from six cohorts of healthy subjects, encompassing 5.2 k proteins, with two novel genome‑wide transcriptomes (57.8 k mRNAs). For 14.8 k protein‑coding transcripts, we assigned the proteins to 21 UniProt‑based classes, based on their preferential intracellular localization and presumed function. This classifed transcriptome‑proteome profle of platelets revealed: (i) Absence of 37.2 k genome‑ wide transcripts. (ii) High quantitative similarity of platelet and megakaryocyte transcriptomes (R = 0.75) for 14.8 k protein‑coding genes, but not for 3.8 k RNA genes or 1.9 k pseudogenes (R = 0.43–0.54), suggesting redistribution of mRNAs upon platelet shedding from megakaryocytes. (iii) Copy numbers of 3.5 k proteins that were restricted in size by the corresponding transcript levels (iv) Near complete coverage of identifed proteins in the relevant transcriptome (log2fpkm > 0.20) except for plasma‑derived secretory proteins, pointing to adhesion and uptake of such proteins. (v) Underrepresentation in the identifed proteome of nuclear‑related, membrane and signaling proteins, as well proteins with low‑level transcripts. -

Induction of Therapeutic Tissue Tolerance Foxp3 Expression Is
Downloaded from http://www.jimmunol.org/ by guest on October 2, 2021 is online at: average * The Journal of Immunology , 13 of which you can access for free at: 2012; 189:3947-3956; Prepublished online 17 from submission to initial decision 4 weeks from acceptance to publication September 2012; doi: 10.4049/jimmunol.1200449 http://www.jimmunol.org/content/189/8/3947 Foxp3 Expression Is Required for the Induction of Therapeutic Tissue Tolerance Frederico S. Regateiro, Ye Chen, Adrian R. Kendal, Robert Hilbrands, Elizabeth Adams, Stephen P. Cobbold, Jianbo Ma, Kristian G. Andersen, Alexander G. Betz, Mindy Zhang, Shruti Madhiwalla, Bruce Roberts, Herman Waldmann, Kathleen F. Nolan and Duncan Howie J Immunol cites 35 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription http://www.jimmunol.org/content/suppl/2012/09/17/jimmunol.120044 9.DC1 This article http://www.jimmunol.org/content/189/8/3947.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2012 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of October 2, 2021. -
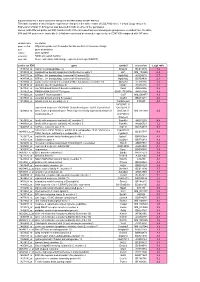
Supplementary File 1
Supplementary file 1: Gene expression changes in the white matter of CD47 KO mice This table contains a list of all gene expression changes in the white matter of CD47 KO mice ( > 2-fold: |Log2 ratio| > 1) Expression of total 14,875 genes was detected in both or either of the genotypes. Genes (with different probe set ID#) found in both of the increased and decreased gene groups were excluded from the table. 594 and 548 genes were markedly (> 2-fold) increased and decreased, respectively, in CD47 KO compared with WT mice. column name description probe set ID# Affymetrix probe set ID number for Mouse 430 2.0 Genome Arrays gene gene description symbol gene symbol accession NCBI accession number Log2 ratio Gene expression fold change expressed as Log2 (KO/WT) probe set ID# gene symbol accession Log2 ratio 1449153_at matrix metallopeptidase 12 Mmp12 BC019135 7.2 1419534_at oxidized low density lipoprotein (lectin-like) receptor 1 Olr1 NM_138648 6.2 1444176_at ATPase, H+ transporting, lysosomal V0 subunit D2 Atp6v0d2 AV204216 5.7 1434798_at ATPase, H+ transporting, lysosomal V0 subunit D2 Atp6v0d2 BB769890 2.3 1420504_at solute carrier family 6 (neurotransmitter transporter), member 14 Slc6a14 AF320226 5.5 1459934_at ubiquitin specific peptidase 42 Usp42 AA516835 5.2 1431800_at von Willebrand factor A domain containing 8 Vwa8 AK004956 5.2 1431927_at RIKEN cDNA 5033417F24 gene 5033417F24Rik AK018199 4.9 1419202_at cystatin F (leukocystatin) Cst7 NM_009977 4.9 1439943_at vacuolar protein sorting 54 (yeast) Vps54 BB201271 4.6 1419966_at tubulin,