Upregulation of Mir-181C Inhibits Chemoresistance by Targeting ST8SIA4 in Chronic Myelocytic Leukemia
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Screening and Identification of Key Biomarkers in Clear Cell Renal Cell Carcinoma Based on Bioinformatics Analysis
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.21.423889; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Screening and identification of key biomarkers in clear cell renal cell carcinoma based on bioinformatics analysis Basavaraj Vastrad1, Chanabasayya Vastrad*2 , Iranna Kotturshetti 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karanataka, India. 3. Department of Ayurveda, Rajiv Gandhi Education Society`s Ayurvedic Medical College, Ron, Karnataka 562209, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2020.12.21.423889; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Clear cell renal cell carcinoma (ccRCC) is one of the most common types of malignancy of the urinary system. The pathogenesis and effective diagnosis of ccRCC have become popular topics for research in the previous decade. In the current study, an integrated bioinformatics analysis was performed to identify core genes associated in ccRCC. An expression dataset (GSE105261) was downloaded from the Gene Expression Omnibus database, and included 26 ccRCC and 9 normal kideny samples. Assessment of the microarray dataset led to the recognition of differentially expressed genes (DEGs), which was subsequently used for pathway and gene ontology (GO) enrichment analysis. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

Aberrant Sialylation in Cancer: Biomarker and Potential Target for Therapeutic Intervention?
cancers Review Aberrant Sialylation in Cancer: Biomarker and Potential Target for Therapeutic Intervention? Silvia Pietrobono * and Barbara Stecca * Tumor Cell Biology Unit, Core Research Laboratory, Institute for Cancer Research and Prevention (ISPRO), Viale Pieraccini 6, 50139 Florence, Italy * Correspondence: [email protected] (S.P.); [email protected] (B.S.); Tel.: +39-055-7944568 (S.P.); +39-055-7944567 (B.S.) Simple Summary: Sialylation is a post-translational modification that consists in the addition of sialic acid to growing glycan chains on glycoproteins and glycolipids. Aberrant sialylation is an established hallmark of several types of cancer, including breast, ovarian, pancreatic, prostate, colorectal and lung cancers, melanoma and hepatocellular carcinoma. Hypersialylation can be the effect of increased activity of sialyltransferases and results in an excess of negatively charged sialic acid on the surface of cancer cells. Sialic acid accumulation contributes to tumor progression by several paths, including stimulation of tumor invasion and migration, and enhancing immune evasion and tumor cell survival. In this review we explore the mechanisms by which sialyltransferases promote cancer progression. In addition, we provide insights into the possible use of sialyltransferases as biomarkers for cancer and summarize findings on the development of sialyltransferase inhibitors as potential anti-cancer treatments. Abstract: Sialylation is an integral part of cellular function, governing many biological processes Citation: Pietrobono, S.; Stecca, B. including cellular recognition, adhesion, molecular trafficking, signal transduction and endocytosis. Aberrant Sialylation in Cancer: Sialylation is controlled by the levels and the activities of sialyltransferases on glycoproteins and Biomarker and Potential Target for lipids. Altered gene expression of these enzymes in cancer yields to cancer-specific alterations of Therapeutic Intervention? Cancers glycoprotein sialylation. -

Downloaded 18 July 2014 with a 1% False Discovery Rate (FDR)
UC Berkeley UC Berkeley Electronic Theses and Dissertations Title Chemical glycoproteomics for identification and discovery of glycoprotein alterations in human cancer Permalink https://escholarship.org/uc/item/0t47b9ws Author Spiciarich, David Publication Date 2017 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California Chemical glycoproteomics for identification and discovery of glycoprotein alterations in human cancer by David Spiciarich A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Chemistry in the Graduate Division of the University of California, Berkeley Committee in charge: Professor Carolyn R. Bertozzi, Co-Chair Professor David E. Wemmer, Co-Chair Professor Matthew B. Francis Professor Amy E. Herr Fall 2017 Chemical glycoproteomics for identification and discovery of glycoprotein alterations in human cancer © 2017 by David Spiciarich Abstract Chemical glycoproteomics for identification and discovery of glycoprotein alterations in human cancer by David Spiciarich Doctor of Philosophy in Chemistry University of California, Berkeley Professor Carolyn R. Bertozzi, Co-Chair Professor David E. Wemmer, Co-Chair Changes in glycosylation have long been appreciated to be part of the cancer phenotype; sialylated glycans are found at elevated levels on many types of cancer and have been implicated in disease progression. However, the specific glycoproteins that contribute to cell surface sialylation are not well characterized, specifically in bona fide human cancer. Metabolic and bioorthogonal labeling methods have previously enabled enrichment and identification of sialoglycoproteins from cultured cells and model organisms. The goal of this work was to develop technologies that can be used for detecting changes in glycoproteins in clinical models of human cancer. -
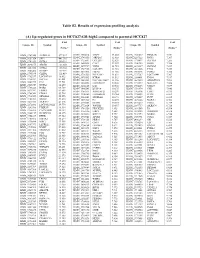
(A) Up-Regulated Genes in HCC827-GR-High2 Compared to Parental HCC827
Table S2. Results of expression profiling analysis (A) Up-regulated genes in HCC827-GR-high2 compared to parental HCC827 Fold Fold Fold Unique ID Symbol Unique ID Symbol Unique ID Symbol change* change* change* ILMN_1709348 ALDH1A1 577.587 ILMN_2310814 MAPT 13.003 ILMN_1741017 PIP4K2B 7.331 ILMN_1651354 SPP1 441.316 ILMN_1748650 MRPL45 12.988 ILMN_3237623 RNY1 7.297 ILMN_1701831 GSTA1 260.591 ILMN_1755897 UGT2B7 12.629 ILMN_1734897 SLC4A4 7.285 ILMN_1658835 CAV2 12.357 ILMN_1746359 RERG 7.280 ILMN_2094875 ABCB1 183.050 ILMN_1678939 VNN2 11.935 ILMN_1671337 SLC2A5 7.257 ILMN_3251540 GSTA2 145.982 ILMN_1729905 GAL3ST1 11.910 ILMN_1691606 LYG2 7.254 ILMN_2062468 IGFBP7 127.721 ILMN_1672536 FBLN1 11.716 ILMN_1785646 PMP22 7.246 ILMN_1795190 CLDN2 111.439 ILMN_1796339 PLEKHA2 11.631 ILMN_1737387 LOC728441 7.207 ILMN_1782937 LOC647169 98.612 ILMN_1676563 HTRA1 11.592 ILMN_1684401 FMO1 7.117 ILMN_1754247 SLC3A1 81.001 ILMN_3263423 LOC100129027 11.346 ILMN_1687035 ADAMTSL4 7.098 ILMN_1662795 CA2 79.581 ILMN_1694898 LOC653857 10.906 ILMN_2153572 MAGEA3 7.086 ILMN_2168747 GSTA2 66.250 ILMN_2404625 LAT 10.560 ILMN_1784283 USH1C 7.079 ILMN_1764228 DAB2 64.709 ILMN_1666546 DUSP14 10.375 ILMN_1731374 CPE 7.046 ILMN_1675797 EPDR1 63.605 ILMN_1764571 ARHGAP23 10.299 ILMN_1765446 EMP3 6.933 ILMN_1708341 PDZK1 59.714 ILMN_3200140 LOC645638 10.284 ILMN_1754002 IL1F8 6.863 ILMN_1713529 SEMA6A 52.575 ILMN_3244343 SNORA21 10.171 ILMN_1878007 FUT9 6.835 ILMN_1708391 NR1H4 43.218 ILMN_1671489 PC 10.075 ILMN_1699208 NAP1L1 6.763 ILMN_2412336 AKR1C2 42.826 ILMN_2404688 -

Supplementary Table 1
Supplementary Table 1. 492 genes are unique to 0 h post-heat timepoint. The name, p-value, fold change, location and family of each gene are indicated. Genes were filtered for an absolute value log2 ration 1.5 and a significance value of p ≤ 0.05. Symbol p-value Log Gene Name Location Family Ratio ABCA13 1.87E-02 3.292 ATP-binding cassette, sub-family unknown transporter A (ABC1), member 13 ABCB1 1.93E-02 −1.819 ATP-binding cassette, sub-family Plasma transporter B (MDR/TAP), member 1 Membrane ABCC3 2.83E-02 2.016 ATP-binding cassette, sub-family Plasma transporter C (CFTR/MRP), member 3 Membrane ABHD6 7.79E-03 −2.717 abhydrolase domain containing 6 Cytoplasm enzyme ACAT1 4.10E-02 3.009 acetyl-CoA acetyltransferase 1 Cytoplasm enzyme ACBD4 2.66E-03 1.722 acyl-CoA binding domain unknown other containing 4 ACSL5 1.86E-02 −2.876 acyl-CoA synthetase long-chain Cytoplasm enzyme family member 5 ADAM23 3.33E-02 −3.008 ADAM metallopeptidase domain Plasma peptidase 23 Membrane ADAM29 5.58E-03 3.463 ADAM metallopeptidase domain Plasma peptidase 29 Membrane ADAMTS17 2.67E-04 3.051 ADAM metallopeptidase with Extracellular other thrombospondin type 1 motif, 17 Space ADCYAP1R1 1.20E-02 1.848 adenylate cyclase activating Plasma G-protein polypeptide 1 (pituitary) receptor Membrane coupled type I receptor ADH6 (includes 4.02E-02 −1.845 alcohol dehydrogenase 6 (class Cytoplasm enzyme EG:130) V) AHSA2 1.54E-04 −1.6 AHA1, activator of heat shock unknown other 90kDa protein ATPase homolog 2 (yeast) AK5 3.32E-02 1.658 adenylate kinase 5 Cytoplasm kinase AK7 -

Translational Profiling Reveals the Transcriptome of Leptin Receptor Neurons and Its Regulation by Leptin
TRANSLATIONAL PROFILING REVEALS THE TRANSCRIPTOME OF LEPTIN RECEPTOR NEURONS AND ITS REGULATION BY LEPTIN by Margaret B. Allison A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Molecular and Integrative Physiology) In the University of Michigan 2015 Doctoral Committee: Professor Martin G. Myers Jr., Chair Associate Professor Carol F. Elias Professor Malcolm J. Low Professor Suzanne Moenter Professor Audrey Seasholtz Before you leave these portals To meet less fortunate mortals There's just one final message I would give to you: You all have learned reliance On the sacred teachings of science So I hope, through life, you never will decline In spite of philistine defiance To do what all good scientists do: Experiment! -- Cole Porter There is no cure for curiosity. -- unknown © Margaret Brewster Allison 2015 ACKNOWLEDGEMENTS If it takes a village to raise a child, it takes a research university to raise a graduate student. There are many people who have supported me over the past six years at Michigan, and it is hard to imagine pursuing my PhD without them. First and foremost among all the people I need to thank is my mentor, Martin. Nothing I might say here would ever suffice to cover the depth and breadth of my gratitude to him. Without his patience, his insight, and his at times insufferably positive outlook, I don’t know where I would be today. Martin supported my intellectual curiosity, honed my scientific inquiry, and allowed me to do some really fun research in his lab. It was a privilege and a pleasure to work for him and with him. -
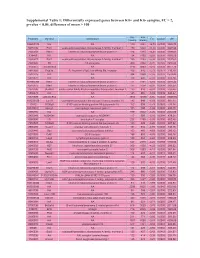
Differentially Expressed Genes Between Kit+ and Kit- Samples, FC
Supplemental Table 1: Differentially expressed genes between Kit+ and Kit- samples, FC > 2, p-value < 0.05, difference of mean > 100 Kit- Kit+ Probeset Symbol Genename FC pvalue diff (mean) (mean) 104280139 NA NA 130 1651 -12,70 0,0000 1520,78 5670239 Ear2 eosinophil-associated, ribonuclease A family, member 2 150 1848 -12,28 0,0000 1697,94 2340358 Ifitm3 interferon induced transmembrane protein 3 126 1315 -10,41 0,0000 1189,02 130465 NA NA 134 1155 -8,60 0,0000 1020,25 2360471 Ear1 eosinophil-associated, ribonuclease A family, member 1 155 1293 -8,34 0,0000 1137,61 7040095 Kit kit oncogene 409 3364 -8,22 0,0145 2955,05 1230347 LOC545854 NA 1216 9893 -8,14 0,0000 8677,76 2810059 Fcgr3a Fc fragment of IgG, low affinity IIIa, receptor 106 816 -7,73 0,0077 710,20 2510725 NA NA 394 2969 -7,53 0,0037 2574,43 5420372 NA NA 128 942 -7,33 0,0000 813,19 101990390 Ifitm2 interferon induced transmembrane protein 2 171 1181 -6,93 0,0000 1010,43 6510075 Ifitm1 interferon induced transmembrane protein 1 152 1054 -6,92 0,0004 901,68 2370286 Slc40a1 solute carrier family 40 (iron-regulated transporter), member 1 133 916 -6,91 0,0000 783,46 5860673 NA NA 143 985 -6,89 0,0239 842,27 6370309 LOC545854 NA 1850 12680 -6,85 0,0000 10829,66 101230129 Ear10 eosinophil-associated, ribonuclease A family, member 10 142 949 -6,68 0,0000 807,11 70112 S100a8 S100 calcium binding protein A8 (calgranulin A) 162 938 -5,79 0,0063 776,16 103780671 Mpeg1 macrophage expressed gene 1 105 598 -5,69 0,0000 493,10 1690184 NA NA 579 3042 -5,26 0,0002 2463,90 2450148 AI324046 expressed -

Regulation Pathway of Mesenchymal Stem Cell Immune Dendritic Cell
Downloaded from http://www.jimmunol.org/ by guest on September 26, 2021 is online at: average * The Journal of Immunology , 13 of which you can access for free at: 2010; 185:5102-5110; Prepublished online 1 from submission to initial decision 4 weeks from acceptance to publication October 2010; doi: 10.4049/jimmunol.1001332 http://www.jimmunol.org/content/185/9/5102 Inhibition of Immune Synapse by Altered Dendritic Cell Actin Distribution: A New Pathway of Mesenchymal Stem Cell Immune Regulation Alessandra Aldinucci, Lisa Rizzetto, Laura Pieri, Daniele Nosi, Paolo Romagnoli, Tiziana Biagioli, Benedetta Mazzanti, Riccardo Saccardi, Luca Beltrame, Luca Massacesi, Duccio Cavalieri and Clara Ballerini J Immunol cites 38 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription http://www.jimmunol.org/content/suppl/2010/10/01/jimmunol.100133 2.DC1 This article http://www.jimmunol.org/content/185/9/5102.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2010 by The American Association of -

Supplemental Figures 04 12 2017
Jung et al. 1 SUPPLEMENTAL FIGURES 2 3 Supplemental Figure 1. Clinical relevance of natural product methyltransferases (NPMTs) in brain disorders. (A) 4 Table summarizing characteristics of 11 NPMTs using data derived from the TCGA GBM and Rembrandt datasets for 5 relative expression levels and survival. In addition, published studies of the 11 NPMTs are summarized. (B) The 1 Jung et al. 6 expression levels of 10 NPMTs in glioblastoma versus non‐tumor brain are displayed in a heatmap, ranked by 7 significance and expression levels. *, p<0.05; **, p<0.01; ***, p<0.001. 8 2 Jung et al. 9 10 Supplemental Figure 2. Anatomical distribution of methyltransferase and metabolic signatures within 11 glioblastomas. The Ivy GAP dataset was downloaded and interrogated by histological structure for NNMT, NAMPT, 12 DNMT mRNA expression and selected gene expression signatures. The results are displayed on a heatmap. The 13 sample size of each histological region as indicated on the figure. 14 3 Jung et al. 15 16 Supplemental Figure 3. Altered expression of nicotinamide and nicotinate metabolism‐related enzymes in 17 glioblastoma. (A) Heatmap (fold change of expression) of whole 25 enzymes in the KEGG nicotinate and 18 nicotinamide metabolism gene set were analyzed in indicated glioblastoma expression datasets with Oncomine. 4 Jung et al. 19 Color bar intensity indicates percentile of fold change in glioblastoma relative to normal brain. (B) Nicotinamide and 20 nicotinate and methionine salvage pathways are displayed with the relative expression levels in glioblastoma 21 specimens in the TCGA GBM dataset indicated. 22 5 Jung et al. 23 24 Supplementary Figure 4. -

Glycobiology of the Epithelial to Mesenchymal Transition
biomedicines Review Glycobiology of the Epithelial to Mesenchymal Transition Michela Pucci, Nadia Malagolini and Fabio Dall’Olio * Department of Experimental, Diagnostic and Specialty Medicine (DIMES), General Pathology Building, University of Bologna, Via San Giacomo 14, 40126 Bologna, Italy; [email protected] (M.P.); [email protected] (N.M.) * Correspondence: [email protected]; Tel.: +39-051-2094704 Abstract: Glycosylation consists in the covalent, enzyme mediated, attachment of sugar chains to proteins and lipids. A large proportion of membrane and secreted proteins are indeed glycopro- teins, while glycolipids are fundamental component of cell membranes. The biosynthesis of sugar chains is mediated by glycosyltransferases, whose level of expression represents a major factor of regulation of the glycosylation process. In cancer, glycosylation undergoes profound changes, which often contribute to invasion and metastasis. Epithelial to mesenchymal transition (EMT) is a key step in metastasis formation and is intimately associated with glycosylation changes. Numerous carbohydrate structures undergo up- or down-regulation during EMT and often regulate the process. In this review, we will discuss the relationship with EMT of the N-glycans, of the different types of O-glycans, including the classical mucin-type, O-GlcNAc, O-linked fucose, O-linked mannose and of glycolipids. Finally, we will discuss the role in EMT of galectins, a major class of mammalian galactoside-binding lectins. While the expression of specific carbohydrate structures can be used as a marker of EMT and of the propensity to migrate, the manipulation of the glycosylation machinery offers new perspectives for cancer treatment through inhibition of EMT. Citation: Pucci, M.; Malagolini, N.; Keywords: glycosylation; glycosyltransferases; galectins; carbohydrate antigens; glycolipids Dall’Olio, F. -

Giantin Knockout Models Reveal a Feedback Loop Between Golgi Function and Glycosyltransferase Expression
bioRxiv preprint doi: https://doi.org/10.1101/123547; this version posted October 19, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 1 Giantin knockout models reveal a feedback loop 2 between Golgi function and glycosyltransferase 3 expression 4 5 Nicola L. Stevenson1, Dylan J. M. Bergen1,2, Roderick E.H. Skinner2, Erika Kague2, Elizabeth 6 Martin-Silverstone2, Kate A. Robson Brown3, Chrissy L. Hammond2, and David J. Stephens1 * 7 8 1 Cell Biology Laboratories, School of Biochemistry, University of Bristol, Biomedical Sciences 9 Building, University Walk, Bristol, UK, BS8 1TD; 10 2 School of Physiology, Pharmacology and Neuroscience, University of Bristol, Biomedical Sciences 11 Building, University Walk, Bristol, UK, BS8 1TD; 12 3 Computed Tomography Laboratory, School of Arts, University of Bristol, 43 Woodland Road, Bristol 13 BS8 1UU 14 * Corresponding author and lead contact: [email protected] 15 Number of words: 5691 16 Summary statement: 17 Knockout of giantin in a genome-engineered cell line and zebrafish models reveals the capacity of 18 the Golgi to control its own biochemistry through changes in gene expression. 1 bioRxiv preprint doi: https://doi.org/10.1101/123547; this version posted October 19, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license.