The Strategy of Fusion Genes Construction Determines Efficient
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Elimination Pathways of Fusion Protein and Peptide Drugs
Review Volume 11, Issue 2, 2021, 9139 - 9147 https://doi.org/10.33263/BRIAC112.91399147 Elimination Pathways of Fusion Protein and Peptide Drugs Ali Khodadoust 1 , Hossein Aghamollaei 2 , Ali Mohammad Latifi 1 , Kazem Hasanpour 3 , Mahdi Kamali 4 , Hamid Tebyanian 5 , Gholamreza Farnoosh 1,* 1 Applied Biotechnology Research Center, Baqiyatallah University of Medical Sciences, Tehran, Iran 2 Chemical Injuries Research Center, Systems biology and Poisonings Institute, Baqiyatallah University of Medical Sciences, Tehran, Iran 3 Sabzevar University of Medical Sciences, School of Medicine, Sabzevar, Iran 4 Nanobiotechnology Research Center, Baqiyatallah University of Medical Sciences, Tehran, Iran 5 Research Center for Prevention of Oral and Dental Diseases, Baqiyatallah University of Medical Sciences, Tehran, Iran * Correspondence: [email protected]; Scopus Author ID 55855454400 Received: 28.07.2020; Revised: 25.08.2020; Accepted: 27.08.2020; Published: 1.09.2020 Abstract: Fusion proteins have been known as an interesting subject for scientific researches in improving properties or making a new function by synergistically incorporating two protein domains into one complex. Fusion proteins are created by ligation of two or more protein domains in a single polypeptide with functional properties. Improvement of therapeutic function is one of the main goals for developing these products. Elimination from the body is one of the most important points that should be considered in the design and production of fusion proteins. This review describes some of the most important excretion/elimination pathways of fusion peptide and protein drugs as well as serious elimination challenges in the development and manufacturing of fusion proteins. Keywords: Fusion proteins; Therapy; Pharmacokinetics; Serum half-life; Peptide. -

CUE-101, a Novel Fc Fusion Protein for Selective Targeting and Expansion of Anti-Tumor T Cells for Treatment of HPV-Driven Malignancies
CUE-101, a novel Fc fusion protein for selective targeting and expansion of anti-tumor T cells for treatment of HPV-driven malignancies Natasha Girgis1, Steven N. Quayle1, Alyssa Nelson1, Dharma Raj Thapa1, Sandrine Hulot1, Lauren Kraemer1, Miguel Moreta1, Zohra Merazga1, Robert Ruidera1, Dominic Beal1, Mark Haydock1, Jonathan Soriano1, Luke Witt1, Jessica Ryabin1, Emily Spaulding1, John F. Ross1, Saso Cemerski1, Anish Suri1, Rodolfo Chaparro1, Ronald Seidel1, Kenneth J. Pienta2, Mary C. Simcox1 1Cue Biopharma, Cambridge, Massachusetts; 2The James Buchanan Brady Urological Institute and the Department of Urology, Johns Hopkins School of Medicine, Baltimore, Maryland + + Background CUE-101 selectively expands E711-20-specific CD8 T mCUE-101 expands functional antigen-specific CD8 T • Human papilloma virus (HPV) is responsible for 72% of oropharyngeal, 90% of cervical, 90% cells from healthy human PBMCs cells in the tumor and the periphery of anal, and 71% of vulvar, vaginal, or penile cancers, causing significant morbidity and A. B. A. B. C. 9 mortality worldwide. Innovative therapies are urgently needed for these malignancies, Vehicle 100 nM CUE-101 107 15 Peripheral Blood particularly in the largely incurable metastatic setting. 106 10 5 • The E7 oncoprotein is constitutively expressed in HPV-associated cancers, is necessary for 105 ⍺PD-1 5 PE 4 Vehicle mCUE-101 Combo - 10 initiation and maintenance of malignant transformation, and is genetically conserved in 1 cancer (Mirabello 2017). 103 1 1 102 E7-Specific of CD8+ T cells % IFNg+,TNFa+ -

Interferon-Based Biopharmaceuticals: Overview on the Production, Purification, and Formulation
Review Interferon-Based Biopharmaceuticals: Overview on the Production, Purification, and Formulation Leonor S. Castro 1,†, Guilherme S. Lobo 1,†, Patrícia Pereira 2 , Mara G. Freire 1 ,Márcia C. Neves 1,* and Augusto Q. Pedro 1,* 1 CICECO–Aveiro Institute of Materials, Chemistry Department, University of Aveiro, Campus Universitário de Santiago, 3810-193 Aveiro, Portugal; [email protected] (L.S.C.); [email protected] (G.S.L.); [email protected] (M.G.F.) 2 Centre for Mechanical Engineering, Materials and Processes, Department of Chemical Engineering, University of Coimbra, Rua Sílvio Lima-Polo II, 3030-790 Coimbra, Portugal; [email protected] * Correspondence: [email protected] (M.C.N.); [email protected] (A.Q.P.) † These authors contributed equally to this work. Abstract: The advent of biopharmaceuticals in modern medicine brought enormous benefits to the treatment of numerous human diseases and improved the well-being of many people worldwide. First introduced in the market in the early 1980s, the number of approved biopharmaceutical products has been steadily increasing, with therapeutic proteins, antibodies, and their derivatives accounting for most of the generated revenues. The success of pharmaceutical biotechnology is closely linked with remarkable developments in DNA recombinant technology, which has enabled the production of proteins with high specificity. Among promising biopharmaceuticals are interferons, first described by Isaacs and Lindenmann in 1957 and approved for clinical use in humans nearly thirty years later. Interferons are secreted autocrine and paracrine proteins, which by regulating several biochemical Citation: Castro, L.S.; Lobo, G.S.; pathways have a spectrum of clinical effectiveness against viral infections, malignant diseases, Pereira, P.; Freire, M.G.; Neves, M.C.; and multiple sclerosis. -
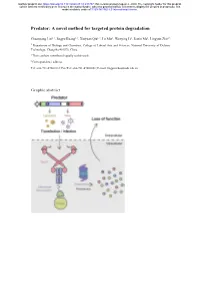
A Novel Method for Targeted Protein Degradation
bioRxiv preprint doi: https://doi.org/10.1101/2020.07.31.231787; this version posted August 2, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Predator: A novel method for targeted protein degradation Chuanyang Liu1, †, Jingyu Kuang1, †, Xinyuan Qiu1, †, Lu Min1, Wenying Li1, Jiaxin Ma1, Lingyun Zhu1,* 1 Department of Biology and Chemistry, College of Liberal Arts and Sciences, National University of Defense Technology, Changsha 410073, China † These authors contributed equally to this work. ∗ Correspondence address. Tel: +86-731-87001812; Fax/Tel: +86-731-87001801; E-mail: [email protected] Graphic abstract bioRxiv preprint doi: https://doi.org/10.1101/2020.07.31.231787; this version posted August 2, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Abstract Protein expression and degradation are fundamental to cell function and physiological status of organisms. Interfering with protein expression not only provides powerful strategies to analyze the function of proteins but also inspires effective treatment methods for diseases caused by protein dysfunction. Recently, harnessing the power of the ubiquitin-proteasome system for targeted protein degradation (TPD) has become the focus of researches. Over the past two decades, TPD technologies, such as E3 ligase modification, PROTACs, and the Trim-Away method, have successfully re-oriented the ubiquitin-proteasome pathway and thus degraded many pathogenic proteins and even "undruggable" targets. -

Overview of Antibody Drug Delivery
pharmaceutics Review Overview of Antibody Drug Delivery Sahar Awwad 1,2,* ID and Ukrit Angkawinitwong 1 1 UCL School of Pharmacy, London WC1N 1AX, UK; [email protected] 2 National Institute for Health Research (NIHR) Biomedical Research Centre at Moorfields Eye Hospital NHS Foundation Trust and UCL Institute of Ophthalmology, London EC1 V9EL, UK * Correspondence: [email protected]; Tel.: +44-207-753-5802 Received: 27 March 2018; Accepted: 29 June 2018; Published: 4 July 2018 Abstract: Monoclonal antibodies (mAbs) are one of the most important classes of therapeutic proteins, which are used to treat a wide number of diseases (e.g., oncology, inflammation and autoimmune diseases). Monoclonal antibody technologies are continuing to evolve to develop medicines with increasingly improved safety profiles, with the identification of new drug targets being one key barrier for new antibody development. There are many opportunities for developing antibody formulations for better patient compliance, cost savings and lifecycle management, e.g., subcutaneous formulations. However, mAb-based medicines also have limitations that impact their clinical use; the most prominent challenges are their short pharmacokinetic properties and stability issues during manufacturing, transport and storage that can lead to aggregation and protein denaturation. The development of long acting protein formulations must maintain protein stability and be able to deliver a large enough dose over a prolonged period. Many strategies are being pursued to improve the formulation and dosage forms of antibodies to improve efficacy and to increase the range of applications for the clinical use of mAbs. Keywords: antibodies; protein; pharmacokinetics; drug delivery; stability 1. -

COVID-19: Mechanisms of Vaccination and Immunity
Review COVID-19: Mechanisms of Vaccination and Immunity Daniel E. Speiser 1,* and Martin F. Bachmann 2,3,4,* 1 Department of Oncology, University Hospital and University of Lausanne, 1066 Lausanne, Switzerland 2 International Immunology Centre, Anhui Agricultural University, Hefei 230036, China 3 Department of Rheumatology, Immunology and Allergology, Inselspital, University of Bern, 3010 Bern, Switzerland 4 Department of BioMedical Research, University of Bern, 3008 Bern, Switzerland * Correspondence: [email protected] (D.E.S.); [email protected] (M.F.B.) Received: 2 July 2020; Accepted: 20 July 2020; Published: 22 July 2020 Abstract: Vaccines are needed to protect from SARS-CoV-2, the virus causing COVID-19. Vaccines that induce large quantities of high affinity virus-neutralizing antibodies may optimally prevent infection and avoid unfavorable effects. Vaccination trials require precise clinical management, complemented with detailed evaluation of safety and immune responses. Here, we review the pros and cons of available vaccine platforms and options to accelerate vaccine development towards the safe immunization of the world’s population against SARS-CoV-2. Favorable vaccines, used in well-designed vaccination strategies, may be critical for limiting harm and promoting trust and a long-term return to normal public life and economy. Keywords: SARS-CoV-2; COVID-19; nucleic acid tests; serology; vaccination; immunity 1. Introduction The COVID-19 pandemic holds great challenges for which the world is only partially prepared [1]. SARS-CoV-2 combines serious pathogenicity with high infectivity. The latter is enhanced by the fact that asymptomatic and pre-symptomatic individuals can transmit the virus, in contrast to SARS-CoV-1 and MERS-CoV, which were transmitted by symptomatic patients and could be contained more efficiently [2,3]. -

Incubation Period of Measles from Exposure to Prodrome ■ Exposure to Rash Onset Averages 11 to 12 Days
Measles Paul Gastanaduy, MD; Penina Haber, MPH; Paul A. Rota, PhD; and Manisha Patel, MD, MS Measles is an acute, viral, infectious disease. References to measles can be found from as early as the 7th century. The Measles disease was described by the Persian physician Rhazes in the ● Acute viral infectious disease 10th century as “more to be dreaded than smallpox.” ● First described in 7th century In 1846, Peter Panum described the incubation period of ● Vaccines first licensed include measles and lifelong immunity after recovery from the disease. measles in 1963, MMR in 1971, John Enders and Thomas Chalmers Peebles isolated the virus in and MMRV in 2005 human and monkey kidney tissue culture in 1954. The first live, ● Infection nearly universal attenuated vaccine (Edmonston B strain) was licensed for use in during childhood in the United States in 1963. In 1971, a combined measles, mumps, prevaccine era and rubella (MMR) vaccine was licensed for use in the United ● Still common and often fatal States. In 2005, a combination measles, mumps, rubella, and in developing countries varicella (MMRV) vaccine was licensed. Before a vaccine was available, infection with measles virus was nearly universal during childhood, and more than 90% of persons were immune due to past infection by age 15 years. Measles is still a common and often fatal disease in developing countries. The World Health Organization estimates there were 142,300 deaths from measles globally in 2018. In the United 13 States, there have been recent outbreaks; the largest occurring in 2019 primarily among people who were not vaccinated. -

Forced Expression of the DEK-NUP214 Fusion Protein
Sandén et al. BMC Cancer 2013, 13:440 http://www.biomedcentral.com/1471-2407/13/440 RESEARCH ARTICLE Open Access Forced expression of the DEK-NUP214 fusion protein promotes proliferation dependent on upregulation of mTOR Carl Sandén1*, Malin Ageberg1, Jessica Petersson1, Andreas Lennartsson2 and Urban Gullberg1 Abstract Background: The t(6;9)(p23;q34) chromosomal translocation is found in 1% of acute myeloid leukemia and encodes the fusion protein DEK-NUP214 (formerly DEK-CAN) with largely uncharacterized functions. Methods: We expressed DEK-NUP214 in the myeloid cell lines U937 and PL-21 and studied the effects on cellular functions. Results: In this study, we demonstrate that expression of DEK-NUP214 increases cellular proliferation. Western blot analysis revealed elevated levels of one of the key proteins regulating proliferation, the mechanistic target of rapamycin, mTOR. This conferred increased mTORC1 but not mTORC2 activity, as determined by the phosphorylation of their substrates, p70 S6 kinase and Akt. The functional importance of the mTOR upregulation was determined by assaying the downstream cellular processes; protein synthesis and glucose metabolism. A global translation assay revealed a substantial increase in the translation rate and a metabolic assay detected a shift from glycolysis to oxidative phosphorylation, as determined by a reduction in lactate production without a concomitant decrease in glucose consumption. Both these effects are in concordance with increased mTORC1 activity. Treatment with the mTORC1 inhibitor everolimus (RAD001) selectively reversed the DEK-NUP214-induced proliferation, demonstrating that the effect is mTOR-dependent. Conclusions: Our study shows that the DEK-NUP214 fusion gene increases proliferation by upregulation of mTOR, suggesting that patients with leukemias carrying DEK-NUP214 may benefit from treatment with mTOR inhibitors. -

Studies to Prevent Degradation of Recombinant Fc-Fusion Protein Expressed in Mammalian Cell Line and Protein Characterization
International Journal of Molecular Sciences Article Studies to Prevent Degradation of Recombinant Fc-Fusion Protein Expressed in Mammalian Cell Line and Protein Characterization Sanjukta Chakrabarti 1,†, Colin J. Barrow 2,†, Rupinder K. Kanwar 3, Venkata Ramana 1,*,† and Jagat R. Kanwar 3,* 1 Reliance Life Sciences, Dhirubhai Ambani Life Sciences Center, Navi Mumbai 400701, India; [email protected] 2 School of Life and Environmental Sciences, Deakin University, Waurn Ponds Campus, Geelong 3216, Australia; [email protected] 3 Nanomedicine-Laboratory of Immunology and Molecular Biomedical Research (NLIMBR), School of Medicine (SoM), Centre for Molecular and Medical Research (C-MMR), Deakin University, Waurn Ponds Campus, Geelong 3216, Australia; [email protected] * Correspondence: [email protected] (V.R.); [email protected] (J.R.K.); Tel.: +91-22-6767-8402 (V.R.); +61-3-5227-1148 (J.R.K.) † These authors contributed equally to this work. Academic Editor: Salvador Ventura Received: 5 January 2016; Accepted: 27 May 2016; Published: 9 June 2016 Abstract: Clipping of recombinant proteins is a major issue in animal cell cultures. A recombinant Fc-fusion protein, VEGFR1(D1–D3)-Fc expressed in CHOK1SV GS-KO cells was observed to be undergoing clippings in lab scale cultures. Partial cleaving of expressed protein initiated early on in cell culture and was observed to increase over time in culture and also on storage. In this study, a few parameters were explored in a bid to inhibit clipping in the fusion protein The effects of culture temperature, duration of culture, the addition of an anti-clumping agent, ferric citrate and use of protease inhibitor cocktail on inhibition of proteolysis of the Fc fusion were studied. -

Experimental Approaches in Delineating Mtor Signaling
G C A T T A C G G C A T genes Review Experimental Approaches in Delineating mTOR Signaling Jiayi Qian 1,2, Siyuan Su 1,2 and Pengda Liu 1,2,* 1 Lineberger Comprehensive Cancer Center, The University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA; [email protected] (J.Q.); [email protected] (S.S.) 2 Department of Biochemistry and Biophysics, The University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA * Correspondence: [email protected]; Tel.: +01-919-966-3522 Received: 11 June 2020; Accepted: 30 June 2020; Published: 2 July 2020 Abstract: The mTOR signaling controls essential biological functions including proliferation, growth, metabolism, autophagy, ageing, and others. Hyperactivation of mTOR signaling leads to a plethora of human disorders; thus, mTOR is an attractive drug target. The discovery of mTOR signaling started from isolation of rapamycin in 1975 and cloning of TOR genes in 1993. In the past 27 years, numerous research groups have contributed significantly to advancing our understanding of mTOR signaling and mTOR biology. Notably, a variety of experimental approaches have been employed in these studies to identify key mTOR pathway members that shape up the mTOR signaling we know today. Technique development drives mTOR research, while canonical biochemical and yeast genetics lay the foundation for mTOR studies. Here in this review, we summarize major experimental approaches used in the past in delineating mTOR signaling, including biochemical immunoprecipitation approaches, genetic approaches, immunofluorescence microscopic approaches, hypothesis-driven studies, protein sequence or motif search driven approaches, and bioinformatic approaches. -

A Chimeric Fusion Protein Containing Transforming Growth Factor-Α
Gene Therapy (1998) 5, 521–530 1998 Stockton Press All rights reserved 0969-7128/98 $12.00 http://www.stockton-press.co.uk/gt A chimeric fusion protein containing transforming growth factor-␣ mediates gene transfer via binding to the EGF receptor J Fominaya1,2, C Uherek1 and W Wels1 1Institute for Experimental Cancer Research, Tumor Biology Center, Breisacher Strasse 117, D-79106 Freiburg, Germany Fusion proteins engineered to incorporate distinct functions binding to EGF receptors. Complexes of the chimeric pro- which co-operate in mediating the cell-type specific uptake tein and plasmid DNA carrying a luciferase reporter gene, and intracellular delivery of DNA present an attractive after condensation with poly-L-lysine resulted in an up to approach for the development of self-assembling vector 150-fold increase in reporter gene expression in EGF systems for targeted gene transfer. Here we have chosen receptor expressing cells in comparison to poly-L-lysine– the EGF receptor overexpressed in many human tumors DNA complexes alone. While in COS-1 cells no of epithelial origin as a target for a novel modular fusion additional endosome escape activity was required, in protein. We have fused a cDNA fragment of the human A431 cells gene delivery was dependent on the simul- EGF receptor ligand TGF-␣ to sequences encoding the taneous presence of the endosome destabilizing translocation domain of Pseudomonas exotoxin A as an reagent chloroquine indicating that cell-type specific fac- endosome escape activity, and the DNA-binding domain of tors such as different intracellular routing of protein–DNA the yeast GAL4 transcription factor. -

Pretreatment with a Monoclonal Antibody/Interleukin-2 Fusion Protein Directed Against DNA Enhances the Delivery of Therapeutic Molecules to Solid Tumors1
Vol. 5, 51–60, January 1999 Clinical Cancer Research 51 Advances in Brief Pretreatment with a Monoclonal Antibody/Interleukin-2 Fusion Protein Directed against DNA Enhances the Delivery of Therapeutic Molecules to Solid Tumors1 Jason L. Hornick, Leslie A. Khawli, Peisheng Hu, 1.7-fold, respectively. Finally, tumor uptake of the radiola- 125 ; Jahangir Sharifi, Chand Khanna, and beled thymidine analogue IUdR also increased 3-fold after pretreatment, indicating that this approach can be Alan L. Epstein2 extended to small molecules such as chemotherapeutic Department of Pathology, University of Southern California School of drugs. Because TNT-3 recognizes a universal nuclear anti- Medicine, Los Angeles, California 90033 [J. L. H., L. A. K., P. H., J. S., A. L. E.] and Pediatric Branch, National Cancer Institute, NIH, gen accessible in degenerating and necrotic cells within all Bethesda, Maryland 20892 [C. K.] solid tumors, this strategy may be applicable to the majority of human cancers. Abstract Introduction The efficacy of molecular therapies for human malig- Physiological barriers to the delivery of therapeutic re- nancies is limited by inadequate accumulation within solid agents to solid tumors are a major obstacle to the clinical tumors. Our laboratory has developed a novel approach that success of developing molecular therapies (1). For example, the uses monoclonal antibodies (MAbs) to direct vasoactive pro- limited clinical responses observed in radioimmunotherapy of teins to tumor sites to increase local vascular permeability solid tumors (2) can be attributed in large part to low tumor and, in turn, improve the delivery of therapeutic reagents. localization of radiolabeled MAb3 (1). Although xenograft mod- Previously, we demonstrated that pretreatment with immu- els in nude mice have shown levels of tumor uptake ranging noconjugates containing interleukin-2 (IL-2) enhances spe- from 1–20% ID/g, patient studies have demonstrated exceed- cific tumor uptake of radiolabeled MAbs without affecting ingly low tumor uptake in the range of 0.01% ID/g (3).