Forced Expression of the DEK-NUP214 Fusion Protein
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Whole-Genome Microarray Detects Deletions and Loss of Heterozygosity of Chromosome 3 Occurring Exclusively in Metastasizing Uveal Melanoma
Anatomy and Pathology Whole-Genome Microarray Detects Deletions and Loss of Heterozygosity of Chromosome 3 Occurring Exclusively in Metastasizing Uveal Melanoma Sarah L. Lake,1 Sarah E. Coupland,1 Azzam F. G. Taktak,2 and Bertil E. Damato3 PURPOSE. To detect deletions and loss of heterozygosity of disease is fatal in 92% of patients within 2 years of diagnosis. chromosome 3 in a rare subset of fatal, disomy 3 uveal mela- Clinical and histopathologic risk factors for UM metastasis noma (UM), undetectable by fluorescence in situ hybridization include large basal tumor diameter (LBD), ciliary body involve- (FISH). ment, epithelioid cytomorphology, extracellular matrix peri- ϩ ETHODS odic acid-Schiff-positive (PAS ) loops, and high mitotic M . Multiplex ligation-dependent probe amplification 3,4 5 (MLPA) with the P027 UM assay was performed on formalin- count. Prescher et al. showed that a nonrandom genetic fixed, paraffin-embedded (FFPE) whole tumor sections from 19 change, monosomy 3, correlates strongly with metastatic death, and the correlation has since been confirmed by several disomy 3 metastasizing UMs. Whole-genome microarray analy- 3,6–10 ses using a single-nucleotide polymorphism microarray (aSNP) groups. Consequently, fluorescence in situ hybridization were performed on frozen tissue samples from four fatal dis- (FISH) detection of chromosome 3 using a centromeric probe omy 3 metastasizing UMs and three disomy 3 tumors with Ͼ5 became routine practice for UM prognostication; however, 5% years’ metastasis-free survival. to 20% of disomy 3 UM patients unexpectedly develop metas- tases.11 Attempts have therefore been made to identify the RESULTS. Two metastasizing UMs that had been classified as minimal region(s) of deletion on chromosome 3.12–15 Despite disomy 3 by FISH analysis of a small tumor sample were found these studies, little progress has been made in defining the key on MLPA analysis to show monosomy 3. -

Meta-Analysis of Nasopharyngeal Carcinoma
BMC Genomics BioMed Central Research article Open Access Meta-analysis of nasopharyngeal carcinoma microarray data explores mechanism of EBV-regulated neoplastic transformation Xia Chen†1,2, Shuang Liang†1, WenLing Zheng1,3, ZhiJun Liao1, Tao Shang1 and WenLi Ma*1 Address: 1Institute of Genetic Engineering, Southern Medical University, Guangzhou, PR China, 2Xiangya Pingkuang associated hospital, Pingxiang, Jiangxi, PR China and 3Southern Genomics Research Center, Guangzhou, Guangdong, PR China Email: Xia Chen - [email protected]; Shuang Liang - [email protected]; WenLing Zheng - [email protected]; ZhiJun Liao - [email protected]; Tao Shang - [email protected]; WenLi Ma* - [email protected] * Corresponding author †Equal contributors Published: 7 July 2008 Received: 16 February 2008 Accepted: 7 July 2008 BMC Genomics 2008, 9:322 doi:10.1186/1471-2164-9-322 This article is available from: http://www.biomedcentral.com/1471-2164/9/322 © 2008 Chen et al; licensee BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Abstract Background: Epstein-Barr virus (EBV) presumably plays an important role in the pathogenesis of nasopharyngeal carcinoma (NPC), but the molecular mechanism of EBV-dependent neoplastic transformation is not well understood. The combination of bioinformatics with evidences from biological experiments paved a new way to gain more insights into the molecular mechanism of cancer. Results: We profiled gene expression using a meta-analysis approach. Two sets of meta-genes were obtained. Meta-A genes were identified by finding those commonly activated/deactivated upon EBV infection/reactivation. -

Genomic Profiling of Adult Acute Lymphoblastic Leukemia by Single
SUPPLEMENTARY APPENDIX Genomic profiling of adult acute lymphoblastic leukemia by single nucleotide polymorphism oligonucleotide microarray and comparison to pediatric acute lymphoblastic leukemia Ryoko Okamoto,1 Seishi Ogawa,2 Daniel Nowak,1 Norihiko Kawamata,1 Tadayuki Akagi,1,3 Motohiro Kato,2 Masashi Sanada,2 Tamara Weiss,4 Claudia Haferlach,4 Martin Dugas,5 Christian Ruckert,5 Torsten Haferlach,4 and H. Phillip Koeffler1,6 1Division of Hematology and Oncology, Cedars-Sinai Medical Center, UCLA School of Medicine, Los Angeles, CA, USA; 2Cancer Genomics Project, Graduate School of Medicine, University of Tokyo, Tokyo, Japan; 3Department of Stem Cell Biology, Graduate School of Medical Science, Kanazawa University 4MLL Munich Leukemia Laboratory, Munich, Germany; 5Department of Medical Informatics and Biomathematics, University of Münster, Münster, Germany; 6Cancer Science Institute of Singapore, National University of Singapore, Singapore Citation: Okamoto R, Ogawa S, Nowak D, Kawamata N, Akagi T, Kato M, Sanada M, Weiss T, Haferlach C, Dugas M, Ruckert C, Haferlach T, and Koeffler HP. Genomic profiling of adult acute lymphoblastic leukemia by single nucleotide polymorphism oligonu- cleotide microarray and comparison to pediatric acute lymphoblastic leukemia. Haematologica 2010;95(9):1481-1488. doi:10.3324/haematol.2009.011114 Online Supplementary Data ed by PCR of genomic DNA and subsequent direct sequencing of SNP in a region of CNN-LOH in an ALL sample versus the corresponding Design and Methods matched normal sample (Online Supplementary -

The DEK Oncoprotein and Its Emerging Roles in Gene Regulation
Leukemia (2015) 29, 1632–1636 © 2015 Macmillan Publishers Limited All rights reserved 0887-6924/15 www.nature.com/leu CONCISE REVIEW The DEK oncoprotein and its emerging roles in gene regulation C Sandén and U Gullberg The DEK oncogene is highly expressed in cells from most human tissues and overexpressed in a large and growing number of cancers. It also fuses with the NUP214 gene to form the DEK-NUP214 fusion gene in a subset of acute myeloid leukemia. Originally characterized as a member of this translocation, DEK has since been implicated in epigenetic and transcriptional regulation, but its role in these processes is still elusive and intriguingly complex. Similarly multifaceted is its contribution to cellular transformation, affecting multiple cellular processes such as self-renewal, proliferation, differentiation, senescence and apoptosis. Recently, the roles of the DEK and DEK-NUP214 proteins have been elucidated by global analysis of DNA binding and gene expression, as well as multiple functional studies. This review outlines recent advances in the understanding of the basic functions of the DEK protein and its role in leukemogenesis. Leukemia (2015) 29, 1632–1636; doi:10.1038/leu.2015.72 INTRODUCTION DNA-binding structure in the C-terminal end of the protein 18 The DEK gene was originally discovered as a fusion partner in the (Figure 1). The specificity of the binding between DEK and DNA (6;9)(p23;q34) chromosomal translocation in acute myeloid has been investigated in several studies, demonstrating that it leukemia (AML), described in detail below.1 Since then, DEK has depends on either the sequence or the structure of the chromatin been shown to be expressed in most human cells and tissues and and that it correlates with the transcriptional activity of the gene. -

Real-Time PCR Analysis of Af4 and Dek Genes Expression in Acute Promyelocytic Leukemia T (15; 17) Patients
EXPERIMENTAL and MOLECULAR MEDICINE, Vol. 36, No. 3, 279-282, June 2004 Real-Time PCR analysis of af4 and dek genes expression in acute promyelocytic leukemia t (15; 17) patients Hakan Savli1, Sema Sirma2, Keywords: af4; APL; dek; Gene expression; Real Time Balint Nagy3, Melih Aktan4, PCR Guncag Dincol4, Zafer Salcioglu5, 4 2,6 Nazan Sarper and Ugur Ozbek Introduction 1 Department of Medical Biology Translocation associated gene fusions are well known Medical Faculty, University of Kocaeli, Kocaeli, Turkey, incidents in acute myeloid leukemia while the other 2Departments of Genetics genetic changes are less known. Acute promyelocytic Institute for Experimental Medicine (DETAE) leukaemia (APL) which is characterized by a recip- Istanbul University, Istanbul, Turkey rocal t (15; 17) translocation of fusing the pml gene 31st Department of Obstetrics and Gynecology to the retinoic acid receptor alpha (rar-alpha) gene, Semmelweis University, Budapest, Hungary but probably there are more oncogenes responsible 4Istanbul Medical Faculty, Istanbul University, Istanbul, Turkey in APL pathogenesis. Among several newly identified 5SSK Bakirkoy Hospital, Istanbul, Turkey oncogenes, dek and af4 are attractive targets for 6Corresponding author: Tel, 90-533-4275272; researchers interested with leukemia. Single role of Fax, 90-212-6311351; E-mail, [email protected] translocation partners dek and af4 genes in leuke- mogenesis have been shown in previous studies Accepted 12 May 2004 (Domer et al., 1993; Larramendy et al., 2002). We also found that dek and af4 genes were down Abbreviations: APL, acute promyelocytic leukaemia regulated during vitamin D dependent differentiation of acute promyelocytic leukaemia cell line HL-60 cells, in our previous studies, using cDNA array technology (Savli et al., 2002). -

Elimination Pathways of Fusion Protein and Peptide Drugs
Review Volume 11, Issue 2, 2021, 9139 - 9147 https://doi.org/10.33263/BRIAC112.91399147 Elimination Pathways of Fusion Protein and Peptide Drugs Ali Khodadoust 1 , Hossein Aghamollaei 2 , Ali Mohammad Latifi 1 , Kazem Hasanpour 3 , Mahdi Kamali 4 , Hamid Tebyanian 5 , Gholamreza Farnoosh 1,* 1 Applied Biotechnology Research Center, Baqiyatallah University of Medical Sciences, Tehran, Iran 2 Chemical Injuries Research Center, Systems biology and Poisonings Institute, Baqiyatallah University of Medical Sciences, Tehran, Iran 3 Sabzevar University of Medical Sciences, School of Medicine, Sabzevar, Iran 4 Nanobiotechnology Research Center, Baqiyatallah University of Medical Sciences, Tehran, Iran 5 Research Center for Prevention of Oral and Dental Diseases, Baqiyatallah University of Medical Sciences, Tehran, Iran * Correspondence: [email protected]; Scopus Author ID 55855454400 Received: 28.07.2020; Revised: 25.08.2020; Accepted: 27.08.2020; Published: 1.09.2020 Abstract: Fusion proteins have been known as an interesting subject for scientific researches in improving properties or making a new function by synergistically incorporating two protein domains into one complex. Fusion proteins are created by ligation of two or more protein domains in a single polypeptide with functional properties. Improvement of therapeutic function is one of the main goals for developing these products. Elimination from the body is one of the most important points that should be considered in the design and production of fusion proteins. This review describes some of the most important excretion/elimination pathways of fusion peptide and protein drugs as well as serious elimination challenges in the development and manufacturing of fusion proteins. Keywords: Fusion proteins; Therapy; Pharmacokinetics; Serum half-life; Peptide. -

CUE-101, a Novel Fc Fusion Protein for Selective Targeting and Expansion of Anti-Tumor T Cells for Treatment of HPV-Driven Malignancies
CUE-101, a novel Fc fusion protein for selective targeting and expansion of anti-tumor T cells for treatment of HPV-driven malignancies Natasha Girgis1, Steven N. Quayle1, Alyssa Nelson1, Dharma Raj Thapa1, Sandrine Hulot1, Lauren Kraemer1, Miguel Moreta1, Zohra Merazga1, Robert Ruidera1, Dominic Beal1, Mark Haydock1, Jonathan Soriano1, Luke Witt1, Jessica Ryabin1, Emily Spaulding1, John F. Ross1, Saso Cemerski1, Anish Suri1, Rodolfo Chaparro1, Ronald Seidel1, Kenneth J. Pienta2, Mary C. Simcox1 1Cue Biopharma, Cambridge, Massachusetts; 2The James Buchanan Brady Urological Institute and the Department of Urology, Johns Hopkins School of Medicine, Baltimore, Maryland + + Background CUE-101 selectively expands E711-20-specific CD8 T mCUE-101 expands functional antigen-specific CD8 T • Human papilloma virus (HPV) is responsible for 72% of oropharyngeal, 90% of cervical, 90% cells from healthy human PBMCs cells in the tumor and the periphery of anal, and 71% of vulvar, vaginal, or penile cancers, causing significant morbidity and A. B. A. B. C. 9 mortality worldwide. Innovative therapies are urgently needed for these malignancies, Vehicle 100 nM CUE-101 107 15 Peripheral Blood particularly in the largely incurable metastatic setting. 106 10 5 • The E7 oncoprotein is constitutively expressed in HPV-associated cancers, is necessary for 105 ⍺PD-1 5 PE 4 Vehicle mCUE-101 Combo - 10 initiation and maintenance of malignant transformation, and is genetically conserved in 1 cancer (Mirabello 2017). 103 1 1 102 E7-Specific of CD8+ T cells % IFNg+,TNFa+ -

The Oncoprotein DEK Affects the Outcome of PARP1/2 Inhibition
bioRxiv preprint doi: https://doi.org/10.1101/555003; this version posted February 19, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. 1 1 The oncoprotein DEK affects the outcome of 2 PARP1/2 inhibition during replication stress 3 Magdalena Ganz1¶, Christopher Vogel1¶, Christina Czada1, Vera Jörke1, Rebecca 4 Kleiner1, Agnieszka Pierzynska-Mach2,3, Francesca Cella Zanacchi2,4, Alberto 5 Diaspro2,5, Ferdinand Kappes6, Alexander Bürkle7, and Elisa Ferrando-May1* 6 1Department of Biology, Bioimaging Center, University of Konstanz, Konstanz, 7 Germany 8 2Nanoscopy and NIC@IIT, Istituto Italiano di Tecnologia, Genoa, Italy 9 3Department of Experimental Oncology, European Institute of Oncology, Milan, Italy 10 4Biophysics Institute (IBF), National Research Council (CNR), Genoa, Italy 11 5DIFILAB, Department of Physics, University of Genoa, Genoa, Italy 12 6Xi’an Jiaotong-Liverpool University, Dushu Lake Higher Education Town, Suzhou, 13 China 14 7Department of Biology, Molecular Toxicology Group, University of Konstanz, 15 Konstanz, Germany 16 17 Short title: DEK and PARP in replication stress 18 19 * Corresponding author 20 email: [email protected] 21 22 ¶These authors contributed equally to this work bioRxiv preprint doi: https://doi.org/10.1101/555003; this version posted February 19, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. -

Functional Annotation of a Full-Length Mouse Cdna Collection
articles Functional annotation of a full-length mouse cDNA collection The RIKEN Genome Exploration Research Group Phase II Team and the FANTOM Consortium* ...............................................................................................................................* A full list of authors appears at the end of the paper ............................................................................................................................................. The RIKEN Mouse Gene Encyclopaedia Project, a systematic approach to determining the full coding potential of the mouse genome, involves collection and sequencing of full-length complementary DNAs and physical mapping of the corresponding genes to the mouse genome. We organized an international functional annotation meeting (FANTOM) to annotate the ®rst 21,076 cDNAs to be analysed in this project. Here we describe the ®rst RIKEN clone collection, which is one of the largest described for any organism. Analysis of these cDNAs extends known gene families and identi®es new ones. In mammals and higher plants, interpreting the genome sequence is Annotation of cDNAs not straightforward: coding regions are interspersed with noncod- An international meeting was held to facilitate functional annota- ing DNA, and an individual gene may give rise to many gene tion of the cDNA sequences. Participants contributed to the devel- products. Thus, genomic sequence cannot be reliably decoded to opment of a web-based annotation interface that should expedite identify the spectrum of messenger RNAs (the transcriptome) and future annotation of additional clones in the Mouse Gene Encyclo- their corresponding protein products (the proteome). This problem paedia project. We agreed on annotation vocabularies and the is illustrated by the different estimates of the number of human application of Gene Ontology (GO) terms (http://genome.gsc. genes (30,000, 35,000 and 120,000)1±3. -

Interferon-Based Biopharmaceuticals: Overview on the Production, Purification, and Formulation
Review Interferon-Based Biopharmaceuticals: Overview on the Production, Purification, and Formulation Leonor S. Castro 1,†, Guilherme S. Lobo 1,†, Patrícia Pereira 2 , Mara G. Freire 1 ,Márcia C. Neves 1,* and Augusto Q. Pedro 1,* 1 CICECO–Aveiro Institute of Materials, Chemistry Department, University of Aveiro, Campus Universitário de Santiago, 3810-193 Aveiro, Portugal; [email protected] (L.S.C.); [email protected] (G.S.L.); [email protected] (M.G.F.) 2 Centre for Mechanical Engineering, Materials and Processes, Department of Chemical Engineering, University of Coimbra, Rua Sílvio Lima-Polo II, 3030-790 Coimbra, Portugal; [email protected] * Correspondence: [email protected] (M.C.N.); [email protected] (A.Q.P.) † These authors contributed equally to this work. Abstract: The advent of biopharmaceuticals in modern medicine brought enormous benefits to the treatment of numerous human diseases and improved the well-being of many people worldwide. First introduced in the market in the early 1980s, the number of approved biopharmaceutical products has been steadily increasing, with therapeutic proteins, antibodies, and their derivatives accounting for most of the generated revenues. The success of pharmaceutical biotechnology is closely linked with remarkable developments in DNA recombinant technology, which has enabled the production of proteins with high specificity. Among promising biopharmaceuticals are interferons, first described by Isaacs and Lindenmann in 1957 and approved for clinical use in humans nearly thirty years later. Interferons are secreted autocrine and paracrine proteins, which by regulating several biochemical Citation: Castro, L.S.; Lobo, G.S.; pathways have a spectrum of clinical effectiveness against viral infections, malignant diseases, Pereira, P.; Freire, M.G.; Neves, M.C.; and multiple sclerosis. -
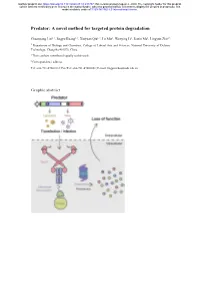
A Novel Method for Targeted Protein Degradation
bioRxiv preprint doi: https://doi.org/10.1101/2020.07.31.231787; this version posted August 2, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Predator: A novel method for targeted protein degradation Chuanyang Liu1, †, Jingyu Kuang1, †, Xinyuan Qiu1, †, Lu Min1, Wenying Li1, Jiaxin Ma1, Lingyun Zhu1,* 1 Department of Biology and Chemistry, College of Liberal Arts and Sciences, National University of Defense Technology, Changsha 410073, China † These authors contributed equally to this work. ∗ Correspondence address. Tel: +86-731-87001812; Fax/Tel: +86-731-87001801; E-mail: [email protected] Graphic abstract bioRxiv preprint doi: https://doi.org/10.1101/2020.07.31.231787; this version posted August 2, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Abstract Protein expression and degradation are fundamental to cell function and physiological status of organisms. Interfering with protein expression not only provides powerful strategies to analyze the function of proteins but also inspires effective treatment methods for diseases caused by protein dysfunction. Recently, harnessing the power of the ubiquitin-proteasome system for targeted protein degradation (TPD) has become the focus of researches. Over the past two decades, TPD technologies, such as E3 ligase modification, PROTACs, and the Trim-Away method, have successfully re-oriented the ubiquitin-proteasome pathway and thus degraded many pathogenic proteins and even "undruggable" targets. -

Overview of Antibody Drug Delivery
pharmaceutics Review Overview of Antibody Drug Delivery Sahar Awwad 1,2,* ID and Ukrit Angkawinitwong 1 1 UCL School of Pharmacy, London WC1N 1AX, UK; [email protected] 2 National Institute for Health Research (NIHR) Biomedical Research Centre at Moorfields Eye Hospital NHS Foundation Trust and UCL Institute of Ophthalmology, London EC1 V9EL, UK * Correspondence: [email protected]; Tel.: +44-207-753-5802 Received: 27 March 2018; Accepted: 29 June 2018; Published: 4 July 2018 Abstract: Monoclonal antibodies (mAbs) are one of the most important classes of therapeutic proteins, which are used to treat a wide number of diseases (e.g., oncology, inflammation and autoimmune diseases). Monoclonal antibody technologies are continuing to evolve to develop medicines with increasingly improved safety profiles, with the identification of new drug targets being one key barrier for new antibody development. There are many opportunities for developing antibody formulations for better patient compliance, cost savings and lifecycle management, e.g., subcutaneous formulations. However, mAb-based medicines also have limitations that impact their clinical use; the most prominent challenges are their short pharmacokinetic properties and stability issues during manufacturing, transport and storage that can lead to aggregation and protein denaturation. The development of long acting protein formulations must maintain protein stability and be able to deliver a large enough dose over a prolonged period. Many strategies are being pursued to improve the formulation and dosage forms of antibodies to improve efficacy and to increase the range of applications for the clinical use of mAbs. Keywords: antibodies; protein; pharmacokinetics; drug delivery; stability 1.