A Chimeric Fusion Protein Containing Transforming Growth Factor-Α
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Elimination Pathways of Fusion Protein and Peptide Drugs
Review Volume 11, Issue 2, 2021, 9139 - 9147 https://doi.org/10.33263/BRIAC112.91399147 Elimination Pathways of Fusion Protein and Peptide Drugs Ali Khodadoust 1 , Hossein Aghamollaei 2 , Ali Mohammad Latifi 1 , Kazem Hasanpour 3 , Mahdi Kamali 4 , Hamid Tebyanian 5 , Gholamreza Farnoosh 1,* 1 Applied Biotechnology Research Center, Baqiyatallah University of Medical Sciences, Tehran, Iran 2 Chemical Injuries Research Center, Systems biology and Poisonings Institute, Baqiyatallah University of Medical Sciences, Tehran, Iran 3 Sabzevar University of Medical Sciences, School of Medicine, Sabzevar, Iran 4 Nanobiotechnology Research Center, Baqiyatallah University of Medical Sciences, Tehran, Iran 5 Research Center for Prevention of Oral and Dental Diseases, Baqiyatallah University of Medical Sciences, Tehran, Iran * Correspondence: [email protected]; Scopus Author ID 55855454400 Received: 28.07.2020; Revised: 25.08.2020; Accepted: 27.08.2020; Published: 1.09.2020 Abstract: Fusion proteins have been known as an interesting subject for scientific researches in improving properties or making a new function by synergistically incorporating two protein domains into one complex. Fusion proteins are created by ligation of two or more protein domains in a single polypeptide with functional properties. Improvement of therapeutic function is one of the main goals for developing these products. Elimination from the body is one of the most important points that should be considered in the design and production of fusion proteins. This review describes some of the most important excretion/elimination pathways of fusion peptide and protein drugs as well as serious elimination challenges in the development and manufacturing of fusion proteins. Keywords: Fusion proteins; Therapy; Pharmacokinetics; Serum half-life; Peptide. -
NUTM1 Is a Recurrent Fusion Gene Partner in B-Cell Precursor Acute
LETTERS TO THE EDITOR However, 20-25% of BCP-ALL patients do not have one NUTM1 is a recurrent fusion gene partner in B-cell of these sentinel cytogenetic aberrations and are there- precursor acute lymphoblastic leukemia associated fore said to have B-other ALL. This B-other ALL subgroup with increased expression of genes on chromosome has an intermediate risk of relapse, but includes both band 10p12.31-12.2 high- and low-risk subgroups that are currently being identified. Our laboratory identified a subtype with a For 20-25% of patients with pediatric B-cell precursor similar expression profile and prognosis as BCR-ABL1, acute lymphoblastic leukemia (BCP-ALL), the driving namely BCR-ABL1-like, within the B-other ALL sub- cytogenetic aberration is unknown. Identification of the group.2 The B-other ALL subgroup also includes other primary lesion could provide better risk stratification and rare cytogenetic subtypes, such as intrachromosomal even identify possible treatment options. We therefore amplification of chromosome 21 and a dicentric chromo- aimed to find novel recurrent genetic aberrations in BCP- 1 ALL cases. We identified an in-frame SLC12A6-NUTM1 some (9;20). It is important to identify more primary fusion, resulting in expression of 3’ exons of NUTM1, lesions in the remaining B-other ALL for better risk strat- and six additional NUTM1-rearranged fusion cases. ification and identification of possible treatment options. These NUTM1-rearranged cases were associated with In this study, we aimed to identify recurrent fusions in high expression of a cluster of genes on chromosome BCP-ALL cases without currently known lesions through band 10p12.31-12.2, including the BMI1 gene. -

CUE-101, a Novel Fc Fusion Protein for Selective Targeting and Expansion of Anti-Tumor T Cells for Treatment of HPV-Driven Malignancies
CUE-101, a novel Fc fusion protein for selective targeting and expansion of anti-tumor T cells for treatment of HPV-driven malignancies Natasha Girgis1, Steven N. Quayle1, Alyssa Nelson1, Dharma Raj Thapa1, Sandrine Hulot1, Lauren Kraemer1, Miguel Moreta1, Zohra Merazga1, Robert Ruidera1, Dominic Beal1, Mark Haydock1, Jonathan Soriano1, Luke Witt1, Jessica Ryabin1, Emily Spaulding1, John F. Ross1, Saso Cemerski1, Anish Suri1, Rodolfo Chaparro1, Ronald Seidel1, Kenneth J. Pienta2, Mary C. Simcox1 1Cue Biopharma, Cambridge, Massachusetts; 2The James Buchanan Brady Urological Institute and the Department of Urology, Johns Hopkins School of Medicine, Baltimore, Maryland + + Background CUE-101 selectively expands E711-20-specific CD8 T mCUE-101 expands functional antigen-specific CD8 T • Human papilloma virus (HPV) is responsible for 72% of oropharyngeal, 90% of cervical, 90% cells from healthy human PBMCs cells in the tumor and the periphery of anal, and 71% of vulvar, vaginal, or penile cancers, causing significant morbidity and A. B. A. B. C. 9 mortality worldwide. Innovative therapies are urgently needed for these malignancies, Vehicle 100 nM CUE-101 107 15 Peripheral Blood particularly in the largely incurable metastatic setting. 106 10 5 • The E7 oncoprotein is constitutively expressed in HPV-associated cancers, is necessary for 105 ⍺PD-1 5 PE 4 Vehicle mCUE-101 Combo - 10 initiation and maintenance of malignant transformation, and is genetically conserved in 1 cancer (Mirabello 2017). 103 1 1 102 E7-Specific of CD8+ T cells % IFNg+,TNFa+ -

Interferon-Based Biopharmaceuticals: Overview on the Production, Purification, and Formulation
Review Interferon-Based Biopharmaceuticals: Overview on the Production, Purification, and Formulation Leonor S. Castro 1,†, Guilherme S. Lobo 1,†, Patrícia Pereira 2 , Mara G. Freire 1 ,Márcia C. Neves 1,* and Augusto Q. Pedro 1,* 1 CICECO–Aveiro Institute of Materials, Chemistry Department, University of Aveiro, Campus Universitário de Santiago, 3810-193 Aveiro, Portugal; [email protected] (L.S.C.); [email protected] (G.S.L.); [email protected] (M.G.F.) 2 Centre for Mechanical Engineering, Materials and Processes, Department of Chemical Engineering, University of Coimbra, Rua Sílvio Lima-Polo II, 3030-790 Coimbra, Portugal; [email protected] * Correspondence: [email protected] (M.C.N.); [email protected] (A.Q.P.) † These authors contributed equally to this work. Abstract: The advent of biopharmaceuticals in modern medicine brought enormous benefits to the treatment of numerous human diseases and improved the well-being of many people worldwide. First introduced in the market in the early 1980s, the number of approved biopharmaceutical products has been steadily increasing, with therapeutic proteins, antibodies, and their derivatives accounting for most of the generated revenues. The success of pharmaceutical biotechnology is closely linked with remarkable developments in DNA recombinant technology, which has enabled the production of proteins with high specificity. Among promising biopharmaceuticals are interferons, first described by Isaacs and Lindenmann in 1957 and approved for clinical use in humans nearly thirty years later. Interferons are secreted autocrine and paracrine proteins, which by regulating several biochemical Citation: Castro, L.S.; Lobo, G.S.; pathways have a spectrum of clinical effectiveness against viral infections, malignant diseases, Pereira, P.; Freire, M.G.; Neves, M.C.; and multiple sclerosis. -
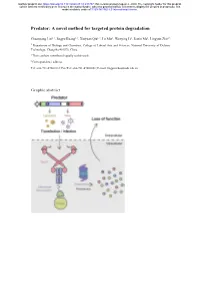
A Novel Method for Targeted Protein Degradation
bioRxiv preprint doi: https://doi.org/10.1101/2020.07.31.231787; this version posted August 2, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Predator: A novel method for targeted protein degradation Chuanyang Liu1, †, Jingyu Kuang1, †, Xinyuan Qiu1, †, Lu Min1, Wenying Li1, Jiaxin Ma1, Lingyun Zhu1,* 1 Department of Biology and Chemistry, College of Liberal Arts and Sciences, National University of Defense Technology, Changsha 410073, China † These authors contributed equally to this work. ∗ Correspondence address. Tel: +86-731-87001812; Fax/Tel: +86-731-87001801; E-mail: [email protected] Graphic abstract bioRxiv preprint doi: https://doi.org/10.1101/2020.07.31.231787; this version posted August 2, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Abstract Protein expression and degradation are fundamental to cell function and physiological status of organisms. Interfering with protein expression not only provides powerful strategies to analyze the function of proteins but also inspires effective treatment methods for diseases caused by protein dysfunction. Recently, harnessing the power of the ubiquitin-proteasome system for targeted protein degradation (TPD) has become the focus of researches. Over the past two decades, TPD technologies, such as E3 ligase modification, PROTACs, and the Trim-Away method, have successfully re-oriented the ubiquitin-proteasome pathway and thus degraded many pathogenic proteins and even "undruggable" targets. -

Transposable Elements in Human Cancers by Genome-Wide EST Alignment
Genes Genet. Syst. (2007) 82, p. 145–156 Transposable elements in human cancers by genome-wide EST alignment Dae-Soo Kim1, Jae-Won Huh2 and Heui-Soo Kim1,2* 1PBBRC, Interdisciplinary Research Program of Bioinformatics, Pusan National University, Busan 609-735, Republic of Korea 2Division of Biological Sciences, College of Natural Sciences, Pusan National University, Busan 609-735, Republic of Korea (Received 24 November 2006, accepted 23 January 2007) Transposable elements may affect coding sequences, splicing patterns, and tran- scriptional regulation of human genes. Particles of the transposable elements have been detected in several tissues and tumors. Here, we report genome-wide analysis of gene expression regulated by transposable elements in human cancers. We adopted an analysis pipeline for screening methods to detect cancer- specific expression from expressed human sequences. We developed a database (TECESdb) for understanding the mechanism of cancer development in relation to transposable elements. A total of 999 genes fused with transposable elements were found to be cancer-related in our analysis of the EST database. According to GO (Gene Ontology) analysis, the majority of the 999 cancer-specific genes have functional association with gene receptor, DNA binding, and kinase activity. Our data could contribute greatly to our understanding of human cancers in relation to transposable elements. Key words: Transposable elements, Cancer, Fusion gene, Bioinformatics, EST also appeared in open-reading frames of functional INTRODUCTION human genes (Yulug et al., 1995; Makalowski et al., 1999; The human genome is estimated to be composed of 45% Nekrutenko and Li, 2001; Huh et al., 2006). transposable elements (International Human Genome The L1 5’UTR element is known to have an antisense Sequencing Consortium 2001). -

Overview of Antibody Drug Delivery
pharmaceutics Review Overview of Antibody Drug Delivery Sahar Awwad 1,2,* ID and Ukrit Angkawinitwong 1 1 UCL School of Pharmacy, London WC1N 1AX, UK; [email protected] 2 National Institute for Health Research (NIHR) Biomedical Research Centre at Moorfields Eye Hospital NHS Foundation Trust and UCL Institute of Ophthalmology, London EC1 V9EL, UK * Correspondence: [email protected]; Tel.: +44-207-753-5802 Received: 27 March 2018; Accepted: 29 June 2018; Published: 4 July 2018 Abstract: Monoclonal antibodies (mAbs) are one of the most important classes of therapeutic proteins, which are used to treat a wide number of diseases (e.g., oncology, inflammation and autoimmune diseases). Monoclonal antibody technologies are continuing to evolve to develop medicines with increasingly improved safety profiles, with the identification of new drug targets being one key barrier for new antibody development. There are many opportunities for developing antibody formulations for better patient compliance, cost savings and lifecycle management, e.g., subcutaneous formulations. However, mAb-based medicines also have limitations that impact their clinical use; the most prominent challenges are their short pharmacokinetic properties and stability issues during manufacturing, transport and storage that can lead to aggregation and protein denaturation. The development of long acting protein formulations must maintain protein stability and be able to deliver a large enough dose over a prolonged period. Many strategies are being pursued to improve the formulation and dosage forms of antibodies to improve efficacy and to increase the range of applications for the clinical use of mAbs. Keywords: antibodies; protein; pharmacokinetics; drug delivery; stability 1. -

EWSR1 Gene EWS RNA Binding Protein 1
EWSR1 gene EWS RNA binding protein 1 Normal Function The EWSR1 gene provides instructions for making the EWS protein, whose function is not completely understood. The EWS protein has two regions that contribute to its function. One region, the transcriptional activation domain, allows the EWS protein to turn on (activate) the first step in the production of proteins from genes (transcription). The other region, the RNA-binding domain, allows the EWS protein to attach (bind) to the genetic blueprint for proteins called RNA. The EWS protein may be involved in piecing together this blueprint. Some studies suggest that the RNA-binding domain is able to block (inhibit) the activity of the transcriptional activation domain, and thus regulate the function of the EWS protein. Health Conditions Related to Genetic Changes Ewing sarcoma Mutations involving the EWSR1 gene can cause a type of cancerous tumor known as Ewing sarcoma. These tumors develop in bones or soft tissues, such as nerves and cartilage. There are several types of Ewing sarcoma, including Ewing sarcoma of bone, extraosseous Ewing sarcoma, peripheral primitive neuroectodermal tumor, and Askin tumor. The mutations that cause these tumors are acquired during a person's lifetime and are present only in the tumor cells. This type of genetic change, called a somatic mutation, is not inherited. The most common mutation that causes Ewing sarcoma is a rearrangement (translocation) of genetic material between chromosome 22 and chromosome 11. This translocation, written as t(11;22), fuses part of the EWSR1 gene on chromosome 22 with part of another gene on chromosome 11 called FLI1, creating an EWSR1/FLI1 fusion gene. -

Birth of a Chimeric Primate Gene by Capture of the Transposase Gene
Birth of a chimeric primate gene by capture of the SEE COMMENTARY transposase gene from a mobile element Richard Cordaux*, Swalpa Udit†, Mark A. Batzer*, and Ce´ dric Feschotte†‡ *Department of Biological Sciences, Biological Computation and Visualization Center, Center for BioModular Multi-Scale Systems, Louisiana State University, 202 Life Sciences Building, Baton Rouge, LA 70803; and †Department of Biology, University of Texas, Arlington, TX 76019 Edited by Susan R. Wessler, University of Georgia, Athens, GA, and approved March 27, 2006 (received for review February 10, 2006) The emergence of new genes and functions is of central impor- SETMAR transcript, which consists of these three exons, is tance to the evolution of species. The contribution of various types predicted to encode a protein of 671 amino acids and is of duplications to genetic innovation has been extensively inves- supported by 48 human cDNA clones from 18 different normal tigated. Less understood is the creation of new genes by recycling and͞or cancerous tissues (Table 1, which is published as sup- of coding material from selfish mobile genetic elements. To inves- porting information on the PNAS web site; refs. 14 and 15). tigate this process, we reconstructed the evolutionary history of These data suggest that the SETMAR protein is broadly ex- SETMAR, a new primate chimeric gene resulting from fusion of a pressed and has an important, yet unknown, function in human. SET histone methyltransferase gene to the transposase gene of a Recently, it was shown that the SET domain of the SETMAR mobile element. We show that the transposase gene was recruited protein exhibits histone methyltransferase activity (15), as do all as part of SETMAR 40–58 million years ago, after the insertion of known SET domains (16, 17). -

Selective Induction of Leukemia-Associated Fusion Genes by High-Dose Ionizing Radiation1
[CANCER RESEARCH 58. 421-425. February I. 1<W8| Selective Induction of Leukemia-associated Fusion Genes by High-Dose Ionizing Radiation1 Michael W. N. Deininger, Shikha Bose, Joanna Gora-Tybor, Xiu-Hua Yan, John M. Goldman, and Junia V. Melo2 Leukaemia Research Fumi Centre for Adult Leukaemia. Department of Haemah>li>/;\: Royal Postgraduale Medical School, Ducane Road, London W12 ONN, United Kingdom ABSTRACT event involves the acquisition of the genetic abnormality whose "success" in the production of a leukemic phenotype will depend on There is strong clinical and epidemiológica! evidence that ionizing its capacity to impart to the target cell a proliferative and/or survival radiation can cause leukemia by inducing DNA damage. This crucial advantage over its normal neighbors. In molecular terms, the gener initiation event is believed to be the result of random DNA breakage and misrepair, whereas the subsequent steps, promotion and progression, ation of a potentially successful reciprocal chromosomal translocation must rely on mechanisms of selective pressure to provide the expanding requires that: (a) at least two independent DNA DSBs occur, one in leukemic population with its proliferative/renewal advantage. To investi each chromosome partner; (b) the two breaks occur simultaneously, gate the susceptibility of human cells to external agents at the genetic i.e., within the same cell cycle, so that the two ends of one broken recombination stage of leukemogenesis, we subjected two hematopoietic chromosome are available to interact and be ligated (misrepaired) to cell lines, KG1 and III.6(1, to high doses of y-irradiation. The irradiation the respective complementary broken ends of the other chromosome; induced the formation of fusion genes characteristic of leukemia in both and (c) the recombination observes the polarity of the DNA molecule. -

DNA Transposons and the Evolution of Eukaryotic Genomes
ANRV329-GE41-15 ARI 12 October 2007 11:1 DNA Transposons and the Evolution of Eukaryotic Genomes Cedric´ Feschotte and Ellen J. Pritham Department of Biology, University of Texas, Arlington, Texas 76019; email: [email protected] Annu. Rev. Genet. 2007. 41:331–68 Key Words The Annual Review of Genetics is online at transposable elements, transposase, molecular domestication, http://genet.annualreviews.org chromosomal rearrangements This article’s doi: 10.1146/annurev.genet.40.110405.090448 Abstract Copyright c 2007 by Annual Reviews. Transposable elements are mobile genetic units that exhibit broad All rights reserved by Fordham University on 11/23/12. For personal use only. diversity in their structure and transposition mechanisms. Transpos- 0066-4197/07/1201-0331$20.00 able elements occupy a large fraction of many eukaryotic genomes and their movement and accumulation represent a major force shap- Annu. Rev. Genet. 2007.41:331-68. Downloaded from www.annualreviews.org ing the genes and genomes of almost all organisms. This review fo- cuses on DNA-mediated or class 2 transposons and emphasizes how this class of elements is distinguished from other types of mobile elements in terms of their structure, amplification dynamics, and genomic effect. We provide an up-to-date outlook on the diversity and taxonomic distribution of all major types of DNA transposons in eukaryotes, including Helitrons and Mavericks. We discuss some of the evolutionary forces that influence their maintenance and di- versification in various genomic environments. Finally, we highlight how the distinctive biological features of DNA transposons have contributed to shape genome architecture and led to the emergence of genetic innovations in different eukaryotic lineages. -

COVID-19: Mechanisms of Vaccination and Immunity
Review COVID-19: Mechanisms of Vaccination and Immunity Daniel E. Speiser 1,* and Martin F. Bachmann 2,3,4,* 1 Department of Oncology, University Hospital and University of Lausanne, 1066 Lausanne, Switzerland 2 International Immunology Centre, Anhui Agricultural University, Hefei 230036, China 3 Department of Rheumatology, Immunology and Allergology, Inselspital, University of Bern, 3010 Bern, Switzerland 4 Department of BioMedical Research, University of Bern, 3008 Bern, Switzerland * Correspondence: [email protected] (D.E.S.); [email protected] (M.F.B.) Received: 2 July 2020; Accepted: 20 July 2020; Published: 22 July 2020 Abstract: Vaccines are needed to protect from SARS-CoV-2, the virus causing COVID-19. Vaccines that induce large quantities of high affinity virus-neutralizing antibodies may optimally prevent infection and avoid unfavorable effects. Vaccination trials require precise clinical management, complemented with detailed evaluation of safety and immune responses. Here, we review the pros and cons of available vaccine platforms and options to accelerate vaccine development towards the safe immunization of the world’s population against SARS-CoV-2. Favorable vaccines, used in well-designed vaccination strategies, may be critical for limiting harm and promoting trust and a long-term return to normal public life and economy. Keywords: SARS-CoV-2; COVID-19; nucleic acid tests; serology; vaccination; immunity 1. Introduction The COVID-19 pandemic holds great challenges for which the world is only partially prepared [1]. SARS-CoV-2 combines serious pathogenicity with high infectivity. The latter is enhanced by the fact that asymptomatic and pre-symptomatic individuals can transmit the virus, in contrast to SARS-CoV-1 and MERS-CoV, which were transmitted by symptomatic patients and could be contained more efficiently [2,3].