Enrichment of Conserved Synaptic Activity-Responsive Element in Neuronal Genes Predicts a Coordinated Response of MEF2, CREB and SRF
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Claire Hastie Thesis
Hastie, Claire E. (2011) Discovering common genetic variants for hypertension using an extreme case-control strategy. PhD thesis. http://theses.gla.ac.uk/2423/ Copyright and moral rights for this thesis are retained by the author A copy can be downloaded for personal non-commercial research or study, without prior permission or charge This thesis cannot be reproduced or quoted extensively from without first obtaining permission in writing from the Author The content must not be changed in any way or sold commercially in any format or medium without the formal permission of the Author When referring to this work, full bibliographic details including the author, title, awarding institution and date of the thesis must be given. Glasgow Theses Service http://theses.gla.ac.uk/ [email protected] Discovering Common Genetic Variants for Hypertension Using an Extreme Case-control Strategy Claire E. Hastie, M.Sc. This being a thesis submitted for the degree of Doctor of Philosophy (Ph.D.) in the Faculty of Medicine, University of Glasgow October 2010 BHF Glasgow Cardiovascular Research Centre Institute of Cardiovascular and Medical Sciences College of Medical, Veterinary, and Life Sciences University of Glasgow © C.E. Hastie 2010 Declaration I declare that this thesis has been written entirely by myself and is a record of research performed by myself with the exception of discovery cohort genotyping (Dr Wai K. Lee, Dr Anna Maria Di Blasio, Stewart Laing, and Dr Davide Gentilini), genotyping and association analysis of replication cohorts (undertaken by investigators from each cohort, respectively), and analysis of data from the BRIGHT (Dr Sandosh Padmanabhan), GRECO and HERCULES clinical functional cohorts (undertaken by investigators from each cohort, respectively). -

Common Differentially Expressed Genes and Pathways Correlating Both Coronary Artery Disease and Atrial Fibrillation
EXCLI Journal 2021;20:126-141– ISSN 1611-2156 Received: December 08, 2020, accepted: January 11, 2021, published: January 18, 2021 Supplementary material to: Original article: COMMON DIFFERENTIALLY EXPRESSED GENES AND PATHWAYS CORRELATING BOTH CORONARY ARTERY DISEASE AND ATRIAL FIBRILLATION Youjing Zheng, Jia-Qiang He* Department of Biomedical Sciences and Pathobiology, College of Veterinary Medicine, Virginia Tech, Blacksburg, VA 24061, USA * Corresponding author: Jia-Qiang He, Department of Biomedical Sciences and Pathobiology, Virginia Tech, Phase II, Room 252B, Blacksburg, VA 24061, USA. Tel: 1-540-231-2032. E-mail: [email protected] https://orcid.org/0000-0002-4825-7046 Youjing Zheng https://orcid.org/0000-0002-0640-5960 Jia-Qiang He http://dx.doi.org/10.17179/excli2020-3262 This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0/). Supplemental Table 1: Abbreviations used in the paper Abbreviation Full name ABCA5 ATP binding cassette subfamily A member 5 ABCB6 ATP binding cassette subfamily B member 6 (Langereis blood group) ABCB9 ATP binding cassette subfamily B member 9 ABCC10 ATP binding cassette subfamily C member 10 ABCC13 ATP binding cassette subfamily C member 13 (pseudogene) ABCC5 ATP binding cassette subfamily C member 5 ABCD3 ATP binding cassette subfamily D member 3 ABCE1 ATP binding cassette subfamily E member 1 ABCG1 ATP binding cassette subfamily G member 1 ABCG4 ATP binding cassette subfamily G member 4 ABHD18 Abhydrolase domain -

Nº Ref Uniprot Proteína Péptidos Identificados Por MS/MS 1 P01024
Document downloaded from http://www.elsevier.es, day 26/09/2021. This copy is for personal use. Any transmission of this document by any media or format is strictly prohibited. Nº Ref Uniprot Proteína Péptidos identificados 1 P01024 CO3_HUMAN Complement C3 OS=Homo sapiens GN=C3 PE=1 SV=2 por 162MS/MS 2 P02751 FINC_HUMAN Fibronectin OS=Homo sapiens GN=FN1 PE=1 SV=4 131 3 P01023 A2MG_HUMAN Alpha-2-macroglobulin OS=Homo sapiens GN=A2M PE=1 SV=3 128 4 P0C0L4 CO4A_HUMAN Complement C4-A OS=Homo sapiens GN=C4A PE=1 SV=1 95 5 P04275 VWF_HUMAN von Willebrand factor OS=Homo sapiens GN=VWF PE=1 SV=4 81 6 P02675 FIBB_HUMAN Fibrinogen beta chain OS=Homo sapiens GN=FGB PE=1 SV=2 78 7 P01031 CO5_HUMAN Complement C5 OS=Homo sapiens GN=C5 PE=1 SV=4 66 8 P02768 ALBU_HUMAN Serum albumin OS=Homo sapiens GN=ALB PE=1 SV=2 66 9 P00450 CERU_HUMAN Ceruloplasmin OS=Homo sapiens GN=CP PE=1 SV=1 64 10 P02671 FIBA_HUMAN Fibrinogen alpha chain OS=Homo sapiens GN=FGA PE=1 SV=2 58 11 P08603 CFAH_HUMAN Complement factor H OS=Homo sapiens GN=CFH PE=1 SV=4 56 12 P02787 TRFE_HUMAN Serotransferrin OS=Homo sapiens GN=TF PE=1 SV=3 54 13 P00747 PLMN_HUMAN Plasminogen OS=Homo sapiens GN=PLG PE=1 SV=2 48 14 P02679 FIBG_HUMAN Fibrinogen gamma chain OS=Homo sapiens GN=FGG PE=1 SV=3 47 15 P01871 IGHM_HUMAN Ig mu chain C region OS=Homo sapiens GN=IGHM PE=1 SV=3 41 16 P04003 C4BPA_HUMAN C4b-binding protein alpha chain OS=Homo sapiens GN=C4BPA PE=1 SV=2 37 17 Q9Y6R7 FCGBP_HUMAN IgGFc-binding protein OS=Homo sapiens GN=FCGBP PE=1 SV=3 30 18 O43866 CD5L_HUMAN CD5 antigen-like OS=Homo -

Sequencing, Analysis, and Annotation of Expressed Sequence Tags for Camelus Dromedarius
University of Nebraska - Lincoln DigitalCommons@University of Nebraska - Lincoln Faculty Publications from the Department of Electrical & Computer Engineering, Department Electrical and Computer Engineering of 2009 Sequencing, Analysis, and Annotation of Expressed Sequence Tags for Camelus dromedarius Abdulaziz M. Al-Swailem King Abdulaziz City for Science and Technology Maher M. Shehata King Abdulaziz City for Science and Technology Faisel M. Abu-Duhier King Abdulaziz City for Science and Technology Essam J. Al-Yamani King Abdulaziz City for Science and Technology Khalid A. Al-Busadah King Faisal University See next page for additional authors Follow this and additional works at: https://digitalcommons.unl.edu/electricalengineeringfacpub Part of the Computer Engineering Commons, and the Electrical and Computer Engineering Commons Al-Swailem, Abdulaziz M.; Shehata, Maher M.; Abu-Duhier, Faisel M.; Al-Yamani, Essam J.; Al-Busadah, Khalid A.; Al-Arawi, Mohammed S.; Al-Khider, Ali Y.; Al-Muhaimeed, Abdullah N.; Al-Qahtani, Fahad H.; Manee, Manee M.; Al-Shomrani, Badr M.; Al-Qhtani, Saad M.; Al-Harthi, Amer S.; Akdemir, Kadir C.; Inan, Mehmet S.; and Otu, Hasan H., "Sequencing, Analysis, and Annotation of Expressed Sequence Tags for Camelus dromedarius" (2009). Faculty Publications from the Department of Electrical and Computer Engineering. 426. https://digitalcommons.unl.edu/electricalengineeringfacpub/426 This Article is brought to you for free and open access by the Electrical & Computer Engineering, Department of at DigitalCommons@University of Nebraska - Lincoln. It has been accepted for inclusion in Faculty Publications from the Department of Electrical and Computer Engineering by an authorized administrator of DigitalCommons@University of Nebraska - Lincoln. Authors Abdulaziz M. Al-Swailem, Maher M. -

Enrichment of Pathogenic Alleles in the Brittle Cornea Gene, ZNF469, in Keratoconus
This is a repository copy of Enrichment of pathogenic alleles in the brittle cornea gene, ZNF469, in keratoconus. White Rose Research Online URL for this paper: http://eprints.whiterose.ac.uk/87454/ Version: Accepted Version Article: Lechner, J, Porter, LF, Rice, A et al. (10 more authors) (2014) Enrichment of pathogenic alleles in the brittle cornea gene, ZNF469, in keratoconus. Human Molecular Genetics, 23 (20). 5527 - 5535. ISSN 0964-6906 https://doi.org/10.1093/hmg/ddu253 Reuse Unless indicated otherwise, fulltext items are protected by copyright with all rights reserved. The copyright exception in section 29 of the Copyright, Designs and Patents Act 1988 allows the making of a single copy solely for the purpose of non-commercial research or private study within the limits of fair dealing. The publisher or other rights-holder may allow further reproduction and re-use of this version - refer to the White Rose Research Online record for this item. Where records identify the publisher as the copyright holder, users can verify any specific terms of use on the publisher’s website. Takedown If you consider content in White Rose Research Online to be in breach of UK law, please notify us by emailing [email protected] including the URL of the record and the reason for the withdrawal request. [email protected] https://eprints.whiterose.ac.uk/ Human Molecular Genetics Enrichment of pathogenic alleles in the brittle cornea gene, ZNF469, in keratoconus Journal: Human Molecular Genetics Manuscript ID: HMG-2014-D-00145.R1 Manuscript ForType: 2 General Peer Article - KReview Office Date Submitted by the Author: n/a Complete List of Authors: ,illoughby. -

Tfcat: the Curated Catalog of Mouse and Human Transcription Factors
Open Access Software2009FultonetVolume al. 10, Issue 3, Article R29 TFCat: the curated catalog of mouse and human transcription factors Debra L Fulton*, Saravanan Sundararajan†, Gwenael Badis‡, Timothy R Hughes‡, Wyeth W Wasserman¤*, Jared C Roach¤§ and Rob Sladek¤† Addresses: *Department of Medical Genetics, Centre for Molecular Medicine and Therapeutics - Child and Family Research Institute, University of British Columbia, West 28th Avenue, Vancouver, V5Z 4H4, Canada. †Departments of Medicine and Human Genetics, McGill University and Genome Quebec Innovation Centre, Dr. Penfield Avenue, Montreal, H3A 1A4, Canada. ‡Banting and Best Department of Medical Research, University of Toronto, College Street, Toronto, M5S 3E1, Canada. §Center for Developmental Therapeutics, Seattle Children's Research Institute, Olive Way, Seattle, 98101, USA. ¤ These authors contributed equally to this work. Correspondence: Rob Sladek. Email: [email protected] Published: 12 March 2009 Received: 5 December 2008 Revised: 26 February 2009 Genome Biology 2009, 10:R29 (doi:10.1186/gb-2009-10-3-r29) Accepted: 12 March 2009 The electronic version of this article is the complete one and can be found online at http://genomebiology.com/2009/10/3/R29 © 2009 Fulton et al.; licensee BioMed Central Ltd. This is an open access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Transcription<p>TFCatof the scientific is a factorcatalog literature</p> catalog of mouse and human transcription factors based on a reliable core collection of annotations obtained by expert review Abstract Unravelling regulatory programs governed by transcription factors (TFs) is fundamental to understanding biological systems. -

Universidade Estadual De Campinas Instituto De Biologia
UNIVERSIDADE ESTADUAL DE CAMPINAS INSTITUTO DE BIOLOGIA CAROLINE BRANDÃO TELES EMPREGANDO A PROTEÔMICA PARA COMPREENDER OS MECANISMOS DE AÇÃO DOS ANTIPSICÓTICOS EM OLIGODENDRÓCITOS HUMANOS EMPLOYING PROTEOMICS TO UNDERSTAND THE MECHANISMS OF ACTION OF ANTIPSYCHOTICS IN HUMAN OLIGODENDROCYTES CAMPINAS 2018 CAROLINE BRANDÃO TELES EMPREGANDO A PROTEÔMICA PARA COMPREENDER OS MECANISMOS DE AÇÃO DOS ANTIPSICÓTICOS EM OLIGODENDRÓCITOS HUMANOS EMPLOYING PROTEOMICS TO UNDERSTAND THE MECHANISMS OF ACTION OF ANTIPSYCHOTICS IN HUMAN OLIGODENDROCYTES Dissertação apresentada ao Instituto de Biologia da Universidade Estadual de Campinas como parte dos requisitos exigidos para a obtenção do título de Mestra em Biologia Funcional e Molecular, na área de Bioquímica Dissertation presented to the Biology Institute of the University of Campinas as part of the requisites required to obtain of Master's degree in Functional and Molecular Biology in the area of Biochemistry ESTE ARQUIVO DIGITAL CORRESPONDE À VERSÃO FINAL DA DISSERTAÇÃO DEFENDIDA PELA ALUNA CAROLINE BRANDÃO TELES, E ORIENTADA PELO PROF. DR. DANIEL MARTINS DE SOUZA E CO- ORIENTADA PELA PROF. DRª JULIANA SILVA CASSOLI Orientador: PROF. DR. DANIEL MARTINS DE SOUZA Coorientador: DRª. JULIANA SILVA CASSOLI CAMPINAS 2018 Agência(s) de fomento e nº(s) de processo(s): FAPESP, 2015/23049-0 Ficha catalográfica Universidade Estadual de Campinas Biblioteca do Instituto de Biologia Mara Janaina de Oliveira - CRB 8/6972 Brandão-Teles, Caroline, 1990- B733e Empregando a proteômica para compreender os mecanismos de ação dos antipsicóticos em oligodendrócitos humanos / Caroline Brandão Teles. – Campinas, SP : [s.n.], 2018. Orientador: Daniel Martins de Souza. Coorientador: Juliana Silva Cassoli. Dissertação (mestrado) – Universidade Estadual de Campinas, Instituto de Biologia. 1. Esquizofrenia. -

Table S1 (Revised)
TABLE 1. LIST OF ALL PROTEINS IDENTIFIED SORTED BY GENE NAME Membrane Probability Accession GI Number Protein Name Entrez Gene Name # of Expts Peptides Fraction score number SV 0.82 10947122 NP_064693.1 ATP-binding cassette, sub-family C, member 9 isoform SUR2B ABCC9 1 EVQM[147]GAVKK SSILIMDEATASIDMATENILQK PM 0.99 21431817 P49597 Protein phosphatase 2C ABI1 (PP2C) (Abscisic acid-insensitive 1) ABI1 1 KEGKDPAAM[147]SAAEYLSK KEGKDPAAMSAAEYLSK PM 0.82 231504 P30172 ACTIN 100,ACTIN 101 AC100 1 AVFPSIVGRPR AGFAGDDAPR VSPDEHPVLLTEAPLNPK SV 1 4501853 NP_001598.1 acetyl-Coenzyme A acyltransferase 1 ACAA1 1 SITVTQDEGIRPSTTMEGLAK PM 0.86 4501855 NP_001084.1 acetyl-Coenzyme A carboxylase beta ACACB 1 VIQVENSHIILTGASALNK M[147]TVPISITNPDLLR MTVPISITNPDLLR EAISNMVVALK SV 0.75 38505218 NP_115545.3 putative acyl-CoA dehydrogenase ACAD11 1 QHSMILVPMNTPGVK QHSM[147]ILVPM[147]NTPGVK PM 1 113018 P08503 Acyl-CoA dehydrogenase, medium-chain specific, mitochondrial precursor (MCAD) ACADM 1 VPKENVLIGEGAGFK PM 0.99 4557237 NP_000010.1 acetyl-Coenzyme A acetyltransferase 1 precursor ACAT1 1 DGLTDVYNK SV 0.89 9998948 NP_064718.1 amiloride-sensitive cation channel 3 isoform c ACCN3 1 DNEETPFEVGIR PM 1 7662238 NP_055792.1 apoptotic chromatin condensation inducer 1 ACIN1 1 VDRPSETKTEEQGIPR CEAEEAEPPAATQPQTSETQTSHL SV 0.76 30089972 NP_004026.2 acyl-coenzyme A oxidase 1 ACOX1 1 EIGTHKPLPGITVGDIGPK TSNHAIVLAQLITK PM 1 16445033 NP_443189.1 ACRC protein; putative nuclear protein ACRC 1 FAKIQIGLKVCDSADR SV 0.93 27477105 NP_055977.3 lipidosin ACSBG1 1 EVEPTSHMGVPR MELPIISNAM[147]LIGDQR -

Sequence Tags for Camelus Dromedarius
Sequencing, Analysis, and Annotation of Expressed Sequence Tags for Camelus dromedarius Abdulaziz M. Al-Swailem1, Maher M. Shehata1, Faisel M. Abu-Duhier1, Essam J. Al-Yamani1, Khalid A. Al-Busadah2, Mohammed S. Al-Arawi1, Ali Y. Al-Khider1, Abdullah N. Al-Muhaimeed1, Fahad H. Al-Qahtani1, Manee M. Manee1, Badr M. Al-Shomrani1, Saad M. Al-Qhtani1, Amer S. Al-Harthi1, Kadir C. Akdemir3, Mehmet S. Inan1{, Hasan H. Otu1,3* 1 Biotechnology Research Center, Natural Resources and Environment Research Institute, King Abdulaziz City for Science and Technology, Riyadh, Saudi Arabia, 2 Faculty of Veterinary Medicine and Animal Resources, King Faisal University, Al-Hassa, Saudi Arabia, 3 Department of Medicine, BIDMC Genomics Center, Harvard Medical School, Boston, Massachusetts, United States of America Abstract Despite its economical, cultural, and biological importance, there has not been a large scale sequencing project to date for Camelus dromedarius. With the goal of sequencing complete DNA of the organism, we first established and sequenced camel EST libraries, generating 70,272 reads. Following trimming, chimera check, repeat masking, cluster and assembly, we obtained 23,602 putative gene sequences, out of which over 4,500 potentially novel or fast evolving gene sequences do not carry any homology to other available genomes. Functional annotation of sequences with similarities in nucleotide and protein databases has been obtained using Gene Ontology classification. Comparison to available full length cDNA sequences and Open Reading Frame (ORF) analysis of camel sequences that exhibit homology to known genes show more than 80% of the contigs with an ORF.300 bp and ,40% hits extending to the start codons of full length cDNAs suggesting successful characterization of camel genes. -
Development of an Osteoblast Cell Culture System for Functional
TECHNISCHE UNIVERSITÄT MÜNCHEN Lehrstuhl für Experimentelle Genetik Development of an osteoblast cell culture system for functional characterization and comparative analyses of mouse models with bone phenotypes and systemic investigation of an Osteogenesis imperfecta disease model Frank Thiele Vollständiger Abdruck der von der Fakultät Wissenschaftszentrum Weihenstephan für Ernährung, Landnutzung und Umwelt der Technischen Universität München zur Erlangung des akademischen Grades eines Doktors der Naturwissenschaften genehmigten Dissertation. Vorsitzender: Univ.-Prof. Dr. A. Gierl Prüfer der Dissertation: 1. Univ.-Prof. Dr. M. Hrabé de Angelis 2. apl. Prof. Dr. J. Adamski Die Dissertation wurde am 24.07.2009 bei der Technischen Universität München eingereicht und durch die Fakultät Wissenschaftszentrum Weihenstephan für Ernährung, Landnutzung und Umwelt am 26.10.2009 angenommen. Für Käte und Erich, Gudrun, Peter, Monique und Alexander I. Table of contents I. TABLE OF CONTENTS............................................................................................ I II. FIGURES AND TABLES ....................................................................................... IV III. ABBREVIATION LIST........................................................................................... VI IV. PUBLICATIONS................................................................................................... IX V. CURRICULUM VITAE........................................................................................... XI VI. AFFIRMATION................................................................................................... -

Membranes of Human Neutrophils Secretory Vesicle Membranes And
Downloaded from http://www.jimmunol.org/ by guest on September 30, 2021 is online at: average * The Journal of Immunology , 25 of which you can access for free at: 2008; 180:5575-5581; ; from submission to initial decision 4 weeks from acceptance to publication J Immunol doi: 10.4049/jimmunol.180.8.5575 http://www.jimmunol.org/content/180/8/5575 Comparison of Proteins Expressed on Secretory Vesicle Membranes and Plasma Membranes of Human Neutrophils Silvia M. Uriarte, David W. Powell, Gregory C. Luerman, Michael L. Merchant, Timothy D. Cummins, Neelakshi R. Jog, Richard A. Ward and Kenneth R. McLeish cites 44 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2008/04/01/180.8.5575.DC1 This article http://www.jimmunol.org/content/180/8/5575.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* • Why • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2008 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 30, 2021. The Journal of Immunology Comparison of Proteins Expressed on Secretory Vesicle Membranes and Plasma Membranes of Human Neutrophils1 Silvia M. -
Supplemental Data and Tables
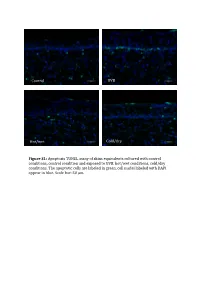
Control UVB Hot/wet Cold/dry Figure S1: Apoptosis TUNEL assay of skins equivalents cultured with control conditions, control condition and exposed to UVB, hot/wet conditions, cold/dry conditions. The apoptotic cells are laBeled in green, cell nuclei laBeled with DAPI appear in blue. Scale bar: 50 μm. Table SI: list of oligonucleotides used in QPCR experiments Gene Forward primer Reverse primer LCE1A TGCAAGAGTGGCTGAGATGC AGACAACACAGTTGGTGT LCE1DE TGAATAGCTGAGAGGTTC CAGCCATGGATCTGCAGAAG LCE5A CTGATGCTAGGTCAGGACTGA TGGTCCTGAGAAGCACTCTAC COL4A1 CCAGGATTTCAAGGTCCAAA CTCCCCTTTGATGATGTCGT LUM TGGCATTGATTGGTGGTACCA TGGGTAGCTTTCAGGGCAGTT FOS CAAGCGGAGACAGACCAACT AGTCAGATCAAGGGAAGCCA Table SII – Part I: genes significantly modulated after treatment 1 (hot-wet) p-value Fold change Gene Symbol Description (Treat-1_vs_CTRL) (Treat-1_vs_CTRL) 0,001147884 15,61495918 MIR639 Homo sapiens microRNA 639 (MIR639), microRNA. 0,021880609 12,6104707 LOC642685 PREDICTED: Homo sapiens similar to maternal G10 transcript (LOC642685), mRNA. 0,010879477 8,858661664 LOC389365 PREDICTED: Homo sapiens hypothetical LOC389365 (LOC389365), mRNA. 0,022200861 6,339924039 HS.566764 DKFZp781A1072_s1 781 (synonym: hlcc4) Homo sapiens cDNA clone DKFZp781A1072 3, mRNA sequence 0,022672172 5,826114515 DKK4 Homo sapiens dickkopf homolog 4 (Xenopus laevis) (DKK4), mRNA. 0,036661342 5,448113208 SLC25A29 Homo sapiens solute carrier family 25, member 29 (SLC25A29), nuclear gene encoding mitochondrial protein, mRNA. 0,035172618 5,141273735 LOC100128361 PREDICTED: Homo sapiens similar to hCG1985411 (LOC100128361), mRNA. 0,01471108 4,783488094 LOC652119 PREDICTED: Homo sapiens similar to putative DUX4 protein (LOC652119), mRNA. 0,009233667 4,511367872 REPS1 Homo sapiens RALBP1 associated Eps domain containing 1 (REPS1), mRNA. 0,031513664 4,407256363 LOC283314 PREDICTED: Homo sapiens misc_RNA (LOC283314), miscRNA. 0,010390588 3,970881226 LOC642773 PREDICTED: Homo sapiens similar to Myc-associated zinc finger protein (MAZI) 0,006806623 3,764373992 LOC391132 PREDICTED: Homo sapiens misc_RNA (LOC391132), miscRNA.