Get High-Res Image
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplementary Table 1.Xlsx
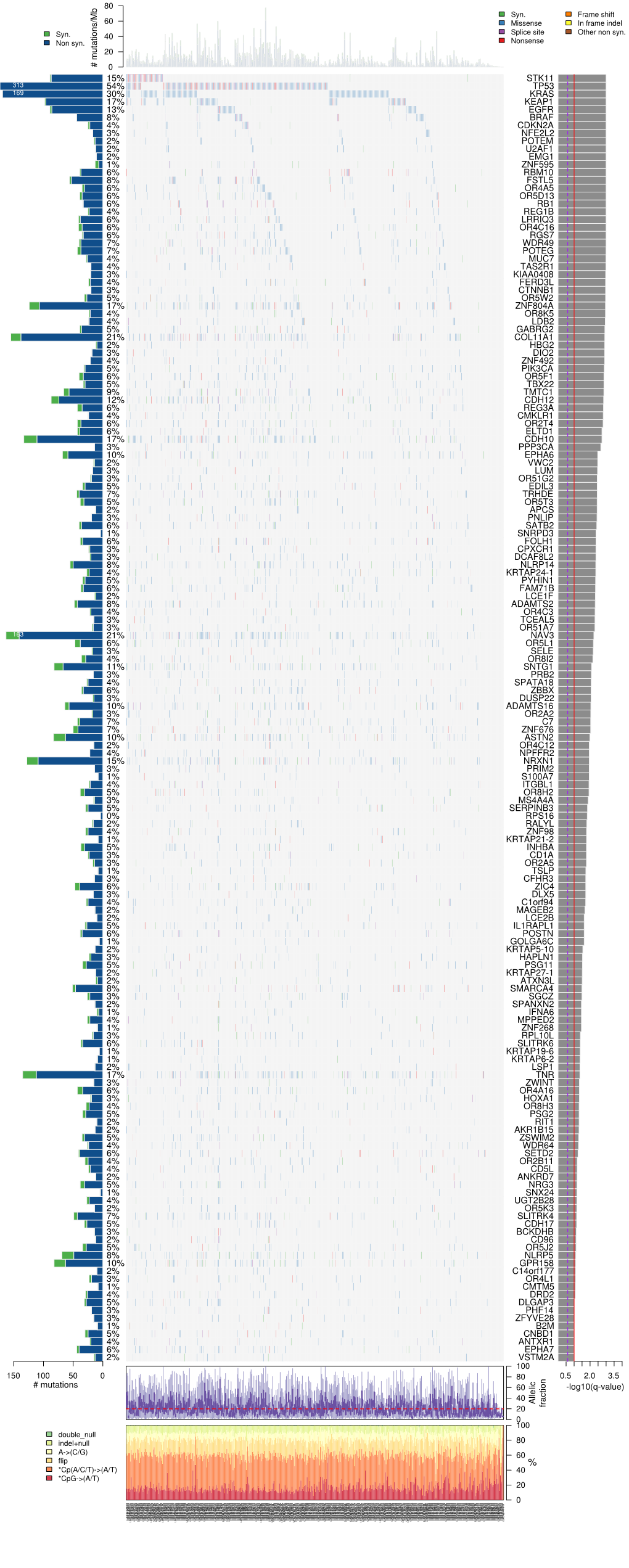
SUPPLEMENTARY APPENDIX Chromothripsis is linked to TP53 alteration, cell cycle impairment, and dismal outcome in acute myeloid leukemia with complex karyotype Frank G. Rücker, 1 Anna Dolnik, 1 Tamara J. Blätte, 1 Veronica Teleanu, 1 Aurélie Ernst, 2 Felicitas Thol, 3 Michael Heuser, 3 Arnold Ganser, 3 Hartmut Döhner, 1 Konstanze Döhner *1 and Lars Bullinger *1,4 1Department of Internal Medicine III, University Hospital of Ulm, Albert-Einstein-Allee 23; 2Division of Molecular Genetics, German Cancer Consor - tium (DKTK), German Cancer Research Center (DKFZ), Heidelberg; 3Department of Hematology, Hemostasis, Oncology, and Stem Cell Transplanta - tion, Hannover Medical School and 4Department of Hematology, Oncology and Tumor Immunology, Charité-University Medicine Berlin, Campus Virchow Klinikum, Berlin, Germany *KD and LB contributed equally to this work. Correspondence: [email protected] doi:10.3324/haematol.2017.180497 Supplementary Table 1 Top 200 differentially expressed genes in chromothripsis‐positive (n=13) vs –negative (n=14) CK‐AML (p‐value <.05) Symbol ID LogFC p value PSMB10 5699 ‐1,652 4,81E‐05 ADAMTS6 11174 ‐1,736 1,50E‐04 SLC7A6 9057 ‐1,317 6,80E‐04 VANGL1 81839 2,403 6,94E‐04 ZNF197 10168 1,409 7,30E‐04 TAS2R62P 338399 ‐1,404 9,22E‐04 PLA2G15 23659 ‐1,455 0,001 CBFB 865 ‐1,453 0,001 CNOT8 9337 ‐1,449 0,001 ACD 65057 ‐1,329 0,001 CKLF 51192 ‐1,657 0,002 CMTM3 123920 ‐1,48 0,002 FAM192A 80011 ‐1,408 0,002 SSH3 54961 ‐1,279 0,002 ZNF415 55786 1,425 0,002 BEX2 84707 1,57 0,002 GCNT2 2651 1,732 0,002 PIM1 5292 1,935 -

Sex-Specific Transcriptome Differences in Human Adipose
G C A T T A C G G C A T genes Article Sex-Specific Transcriptome Differences in Human Adipose Mesenchymal Stem Cells 1, 2, 3 1,3 Eva Bianconi y, Raffaella Casadei y , Flavia Frabetti , Carlo Ventura , Federica Facchin 1,3,* and Silvia Canaider 1,3 1 National Laboratory of Molecular Biology and Stem Cell Bioengineering of the National Institute of Biostructures and Biosystems (NIBB)—Eldor Lab, at the Innovation Accelerator, CNR, Via Piero Gobetti 101, 40129 Bologna, Italy; [email protected] (E.B.); [email protected] (C.V.); [email protected] (S.C.) 2 Department for Life Quality Studies (QuVi), University of Bologna, Corso D’Augusto 237, 47921 Rimini, Italy; [email protected] 3 Department of Experimental, Diagnostic and Specialty Medicine (DIMES), University of Bologna, Via Massarenti 9, 40138 Bologna, Italy; fl[email protected] * Correspondence: [email protected]; Tel.: +39-051-2094114 These authors contributed equally to this work. y Received: 1 July 2020; Accepted: 6 August 2020; Published: 8 August 2020 Abstract: In humans, sexual dimorphism can manifest in many ways and it is widely studied in several knowledge fields. It is increasing the evidence that also cells differ according to sex, a correlation still little studied and poorly considered when cells are used in scientific research. Specifically, our interest is on the sex-related dimorphism on the human mesenchymal stem cells (hMSCs) transcriptome. A systematic meta-analysis of hMSC microarrays was performed by using the Transcriptome Mapper (TRAM) software. This bioinformatic tool was used to integrate and normalize datasets from multiple sources and allowed us to highlight chromosomal segments and genes differently expressed in hMSCs derived from adipose tissue (hADSCs) of male and female donors. -

WO 2019/068007 Al Figure 2
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization I International Bureau (10) International Publication Number (43) International Publication Date WO 2019/068007 Al 04 April 2019 (04.04.2019) W 1P O PCT (51) International Patent Classification: (72) Inventors; and C12N 15/10 (2006.01) C07K 16/28 (2006.01) (71) Applicants: GROSS, Gideon [EVIL]; IE-1-5 Address C12N 5/10 (2006.0 1) C12Q 1/6809 (20 18.0 1) M.P. Korazim, 1292200 Moshav Almagor (IL). GIBSON, C07K 14/705 (2006.01) A61P 35/00 (2006.01) Will [US/US]; c/o ImmPACT-Bio Ltd., 2 Ilian Ramon St., C07K 14/725 (2006.01) P.O. Box 4044, 7403635 Ness Ziona (TL). DAHARY, Dvir [EilL]; c/o ImmPACT-Bio Ltd., 2 Ilian Ramon St., P.O. (21) International Application Number: Box 4044, 7403635 Ness Ziona (IL). BEIMAN, Merav PCT/US2018/053583 [EilL]; c/o ImmPACT-Bio Ltd., 2 Ilian Ramon St., P.O. (22) International Filing Date: Box 4044, 7403635 Ness Ziona (E.). 28 September 2018 (28.09.2018) (74) Agent: MACDOUGALL, Christina, A. et al; Morgan, (25) Filing Language: English Lewis & Bockius LLP, One Market, Spear Tower, SanFran- cisco, CA 94105 (US). (26) Publication Language: English (81) Designated States (unless otherwise indicated, for every (30) Priority Data: kind of national protection available): AE, AG, AL, AM, 62/564,454 28 September 2017 (28.09.2017) US AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, BZ, 62/649,429 28 March 2018 (28.03.2018) US CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, (71) Applicant: IMMP ACT-BIO LTD. -

Sean Raspet – Molecules
1. Commercial name: Fructaplex© IUPAC Name: 2-(3,3-dimethylcyclohexyl)-2,5,5-trimethyl-1,3-dioxane SMILES: CC1(C)CCCC(C1)C2(C)OCC(C)(C)CO2 Molecular weight: 240.39 g/mol Volume (cubic Angstroems): 258.88 Atoms number (non-hydrogen): 17 miLogP: 4.43 Structure: Biological Properties: Predicted Druglikenessi: GPCR ligand -0.23 Ion channel modulator -0.03 Kinase inhibitor -0.6 Nuclear receptor ligand 0.15 Protease inhibitor -0.28 Enzyme inhibitor 0.15 Commercial name: Fructaplex© IUPAC Name: 2-(3,3-dimethylcyclohexyl)-2,5,5-trimethyl-1,3-dioxane SMILES: CC1(C)CCCC(C1)C2(C)OCC(C)(C)CO2 Predicted Olfactory Receptor Activityii: OR2L13 83.715% OR1G1 82.761% OR10J5 80.569% OR2W1 78.180% OR7A2 77.696% 2. Commercial name: Sylvoxime© IUPAC Name: N-[4-(1-ethoxyethenyl)-3,3,5,5tetramethylcyclohexylidene]hydroxylamine SMILES: CCOC(=C)C1C(C)(C)CC(CC1(C)C)=NO Molecular weight: 239.36 Volume (cubic Angstroems): 252.83 Atoms number (non-hydrogen): 17 miLogP: 4.33 Structure: Biological Properties: Predicted Druglikeness: GPCR ligand -0.6 Ion channel modulator -0.41 Kinase inhibitor -0.93 Nuclear receptor ligand -0.17 Protease inhibitor -0.39 Enzyme inhibitor 0.01 Commercial name: Sylvoxime© IUPAC Name: N-[4-(1-ethoxyethenyl)-3,3,5,5tetramethylcyclohexylidene]hydroxylamine SMILES: CCOC(=C)C1C(C)(C)CC(CC1(C)C)=NO Predicted Olfactory Receptor Activity: OR52D1 71.900% OR1G1 70.394% 0R52I2 70.392% OR52I1 70.390% OR2Y1 70.378% 3. Commercial name: Hyperflor© IUPAC Name: 2-benzyl-1,3-dioxan-5-one SMILES: O=C1COC(CC2=CC=CC=C2)OC1 Molecular weight: 192.21 g/mol Volume -

Clinical, Molecular, and Immune Analysis of Dabrafenib-Trametinib
Supplementary Online Content Chen G, McQuade JL, Panka DJ, et al. Clinical, molecular and immune analysis of dabrafenib-trametinib combination treatment for metastatic melanoma that progressed during BRAF inhibitor monotherapy: a phase 2 clinical trial. JAMA Oncology. Published online April 28, 2016. doi:10.1001/jamaoncol.2016.0509. eMethods. eReferences. eTable 1. Clinical efficacy eTable 2. Adverse events eTable 3. Correlation of baseline patient characteristics with treatment outcomes eTable 4. Patient responses and baseline IHC results eFigure 1. Kaplan-Meier analysis of overall survival eFigure 2. Correlation between IHC and RNAseq results eFigure 3. pPRAS40 expression and PFS eFigure 4. Baseline and treatment-induced changes in immune infiltrates eFigure 5. PD-L1 expression eTable 5. Nonsynonymous mutations detected by WES in baseline tumors This supplementary material has been provided by the authors to give readers additional information about their work. © 2016 American Medical Association. All rights reserved. Downloaded From: https://jamanetwork.com/ on 09/30/2021 eMethods Whole exome sequencing Whole exome capture libraries for both tumor and normal samples were constructed using 100ng genomic DNA input and following the protocol as described by Fisher et al.,3 with the following adapter modification: Illumina paired end adapters were replaced with palindromic forked adapters with unique 8 base index sequences embedded within the adapter. In-solution hybrid selection was performed using the Illumina Rapid Capture Exome enrichment kit with 38Mb target territory (29Mb baited). The targeted region includes 98.3% of the intervals in the Refseq exome database. Dual-indexed libraries were pooled into groups of up to 96 samples prior to hybridization. -

An Analysis About Heterogeneity Among Cancers Based on the DNA Methylation Patterns
An analysis about heterogeneity among cancers based on the DNA Methylation Patterns Yang Liu Harbin Institute of Technology Yue Gu Harbin Medical University Mu Su Harbin Institute of Technology Hui Liu Harbin Institute of Technology Shumei Zhang Harbin Medical University Yan Zhang ( [email protected] ) Harbin Institute of Technology https://orcid.org/0000-0002-5307-2484 Research article Keywords: DNA methylation, cancer, epigenetic heterogeneity, survival analysis Posted Date: July 17th, 2019 DOI: https://doi.org/10.21203/rs.2.11636/v1 License: This work is licensed under a Creative Commons Attribution 4.0 International License. Read Full License Version of Record: A version of this preprint was published at BMC Cancer on December 1st, 2019. See the published version at https://doi.org/10.1186/s12885-019-6455-x. Page 1/28 Abstract Background: The occurrence of cancer is usually the result of a co-effect of genetic and environmental factors. It is generally believed that the main cause of cancer is the accumulation of genetic mutations, and DNA methylation, as one of the epigenetic modications closely related to environmental factors, participates in the regulation of gene expression and cell differentiation and plays an important role in the development of cancer. Methods: This article discusses the epigenetic heterogeneity of cancer in detail. Firstly DNA methylation data of 7 cancer types were obtained from Illumina Innium HumanMethylation 450K platform of TCGA database. Diagnostic markers of each cancer were obtained by t-test and absolute difference of DNA differencial methylation analysis. Enrichment analysis of these specic markers indicated that they were involved in different biological functions. -

Figure S1. Effects of Mir‑95 on the Proliferation, Cell Cycle Progression and Apoptosis of the OS Saos‑2 Cell Line
Figure S1. Effects of miR‑95 on the proliferation, cell cycle progression and apoptosis of the OS Saos‑2 cell line. (A and B) The miR‑95 inhibitor significantly inhibited the proliferation and cell cycle progression of Saos‑2 cells, as shown by EdU and PI staining. (C and D) The miR‑95 inhibitor significantly promoted apoptosis of Saos‑2 cells, as determined via PI/Annexin V and TUNEL staining. *P<0.05, **P<0.01, compared with the NC group. NC, negative control; OS, osteosarcoma. Figure S2. SCNN1A is a critical target during miR‑95‑induced OS cell apoptosis. (A) OS Saos‑2 cells were transfected with the miR‑95 inhibitor and shSCNN1A (Sh1 SCNN1A and Sh2 SCNN1A). Real‑time PCR was performed to examine the level of SCNN1A. (B) Cell viability was evaluated via the CCK‑8 assay in the transfected Saos‑2 cells. (C and D) Cell cycle distribution and apoptosis were measured via PI and PI/Annexin V assays in the transfected Saos‑2 cells. *P<0.05, **P<0.01, compared with the NC group. NC, negative control; SC, scramble control; SCNN1A, sodium channel epithelial 1α subunit; OS, osteosarcoma. Table SI. Potential target genes (n=52) of miR‑95 predicted by TargetScan. Gene Cumulative weighted context++ score Total context++ score QDPR ‑0.36 ‑0.45 MGLL ‑0.2 ‑0.37 RAI14 ‑0.45 ‑0.47 ZFHX3 ‑0.2 ‑0.2 KIF4B ‑0.57 ‑0.57 ARPP19 ‑0.56 ‑0.56 TMEM33 ‑0.19 ‑0.4 FOXK1 ‑0.17 ‑0.21 ZNF708 ‑0.16 ‑0.32 ZNF131 ‑0.58 ‑0.58 KIAA0226L ‑0.1 ‑0.33 LGI2 ‑0.19 ‑0.19 C9orf163 ‑0.57 ‑0.57 SNX1 ‑0.3 ‑0.45 CDON ‑0.19 ‑0.19 PLXNA4 ‑0.1 ‑0.18 FAM212B ‑0.5 ‑0.5 ADH5 ‑0.17 ‑0.57 NTSR1 -

The Hypothalamus As a Hub for SARS-Cov-2 Brain Infection and Pathogenesis
bioRxiv preprint doi: https://doi.org/10.1101/2020.06.08.139329; this version posted June 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. The hypothalamus as a hub for SARS-CoV-2 brain infection and pathogenesis Sreekala Nampoothiri1,2#, Florent Sauve1,2#, Gaëtan Ternier1,2ƒ, Daniela Fernandois1,2 ƒ, Caio Coelho1,2, Monica ImBernon1,2, Eleonora Deligia1,2, Romain PerBet1, Vincent Florent1,2,3, Marc Baroncini1,2, Florence Pasquier1,4, François Trottein5, Claude-Alain Maurage1,2, Virginie Mattot1,2‡, Paolo GiacoBini1,2‡, S. Rasika1,2‡*, Vincent Prevot1,2‡* 1 Univ. Lille, Inserm, CHU Lille, Lille Neuroscience & Cognition, DistAlz, UMR-S 1172, Lille, France 2 LaBoratorY of Development and PlasticitY of the Neuroendocrine Brain, FHU 1000 daYs for health, EGID, School of Medicine, Lille, France 3 Nutrition, Arras General Hospital, Arras, France 4 Centre mémoire ressources et recherche, CHU Lille, LiCEND, Lille, France 5 Univ. Lille, CNRS, INSERM, CHU Lille, Institut Pasteur de Lille, U1019 - UMR 8204 - CIIL - Center for Infection and ImmunitY of Lille (CIIL), Lille, France. # and ƒ These authors contriButed equallY to this work. ‡ These authors directed this work *Correspondence to: [email protected] and [email protected] Short title: Covid-19: the hypothalamic hypothesis 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.06.08.139329; this version posted June 19, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

Supplementary Table 1
Supplementary Table 1. List of genes that encode proteins contianing cell surface epitopes and are represented on Agilent human 1A V2 microarray chip (2,177 genes) Agilent Probe ID Gene Symbol GenBank ID UniGene ID A_23_P103803 FCRH3 AF459027 Hs.292449 A_23_P104811 TREH AB000824 Hs.129712 A_23_P105100 IFITM2 X57351 Hs.174195 A_23_P107036 C17orf35 X51804 Hs.514009 A_23_P110736 C9 BC020721 Hs.1290 A_23_P111826 SPAM1 NM_003117 Hs.121494 A_23_P119533 EFNA2 AJ007292 No-Data A_23_P120105 KCNS3 BC004987 Hs.414489 A_23_P128195 HEM1 NM_005337 Hs.182014 A_23_P129332 PKD1L2 BC014157 Hs.413525 A_23_P130203 SYNGR2 AJ002308 Hs.464210 A_23_P132700 TDGF1 X14253 Hs.385870 A_23_P1331 COL13A1 NM_005203 Hs.211933 A_23_P138125 TOSO BC006401 Hs.58831 A_23_P142830 PLA2R1 U17033 Hs.410477 A_23_P146506 GOLPH2 AF236056 Hs.494337 A_23_P149569 MG29 No-Data No-Data A_23_P150590 SLC22A9 NM_080866 Hs.502772 A_23_P151166 MGC15619 BC009731 Hs.334637 A_23_P152620 TNFSF13 NM_172089 Hs.54673 A_23_P153986 KCNJ3 U39196 No-Data A_23_P154855 KCNE1 NM_000219 Hs.121495 A_23_P157380 KCTD7 AK056631 Hs.520914 A_23_P157428 SLC10A5 AK095808 Hs.531449 A_23_P160159 SLC2A5 BC001820 Hs.530003 A_23_P162162 KCTD14 NM_023930 Hs.17296 A_23_P162668 CPM BC022276 Hs.334873 A_23_P164341 VAMP2 AL050223 Hs.25348 A_23_P165394 SLC30A6 NM_017964 Hs.552598 A_23_P167276 PAQR3 AK055774 Hs.368305 A_23_P170636 KCNH8 AY053503 Hs.475656 A_23_P170736 MMP17 NM_016155 Hs.159581 A_23_P170959 LMLN NM_033029 Hs.518540 A_23_P19042 GABRB2 NM_021911 Hs.87083 A_23_P200361 CLCN6 X83378 Hs.193043 A_23_P200710 PIK3C2B -

Supplementary Table S4
Supplementary Table 4 - Patient P10 Heat maps of mutations and gene expression for patients with three or more tumor sample analyzed. P10 SNV Allele frequency Log2(FPKM+1) GeneSymbol S1 S5 S8 S1 S5 S8 S1 S5 S8 Ref KDM6A* 1 1 1 0.82 0.87 0.64 2.36 2.28 2.22 3.01 UBAP2L 1 1 1 0.63 0.58 0.47 4.72 5.10 5.33 4.78 SLC12A5 1 1 1 0.60 0.32 0.29 0.05 0.01 0.00 0.02 KDM6A FMO1 1 1 1 0.59 0.56 0.41 0.23 0.04 0.40 0.14 MCC 1 1 1 0.58 0.46 0.49 4.33 5.29 4.99 4.54 3.50 MCAM 1 1 1 0.57 0.38 0.38 2.73 2.60 3.95 3.11 CCDC60 1 1 1 0.52 0.47 0.36 0.06 0.01 0.00 1.58 3.00 RALYL 1 1 1 0.52 0.25 0.24 0.03 0.03 0.00 0.15 2.50 AGXT 1 1 1 0.50 0.40 0.29 0.01 0.00 0.00 0.04 FAM209B 1 1 1 0.46 0.33 0.35 0.01 0.02 0.02 0.09 2.00 MYO7B 1 1 1 0.45 0.52 0.34 0.01 0.00 0.01 0.13 PPARG 1 1 1 0.45 0.48 0.34 5.55 5.93 5.70 5.92 1.50 MUC4 1 1 1 0.44 0.42 0.29 0.17 0.12 0.05 0.67 Log2(FPKM+1) HRAS** 1 1 1 0.44 0.39 0.20 4.32 4.75 4.55 3.28 1.00 MIA3 1 1 1 0.39 0.38 0.19 5.30 5.35 5.15 4.70 0.50 CLTC# 1 1 1 0.39 0.39 0.37 5.57 5.93 5.81 6.05 CEP152 1 1 1 0.38 0.43 0.17 3.28 2.78 3.25 2.14 0.00 DDC 1 1 1 0.38 0.34 0.29 0.01 0.01 0.00 0.12 S1 S5 S8 Ref HERC2 1 1 1 0.38 0.36 0.25 3.60 3.77 3.09 3.14 SEMA3D 1 1 1 0.37 0.51 0.26 0.38 0.60 0.53 2.83 ZNF320 1 1 1 0.34 0.41 0.22 4.14 4.25 4.08 4.07 HRAS ERBB3# 1 1 1 0.34 0.39 0.26 5.52 5.85 5.60 4.69 KSR2 1 1 1 0.33 0.39 0.27 0.75 1.17 0.65 0.97 5.00 TRERF1 1 1 1 0.32 0.42 0.27 2.10 2.45 2.25 2.63 4.50 COG4 1 1 1 0.31 0.36 0.27 3.78 4.06 4.12 3.58 4.00 LPAR5 1 1 1 0.31 0.43 0.24 0.92 1.03 0.47 1.17 3.50 PRTN3 1 1 1 0.30 0.05 0.02 -

Reproducible Copy Number Variation Patterns Among Single Circulating Tumor Cells of Lung Cancer Patients
Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients Xiaohui Nia,b,1, Minglei Zhuoc,1, Zhe Sua,1, Jianchun Duanc,1, Yan Gaoa,1, Zhijie Wangc,1, Chenghang Zongb,1,2, Hua Baic, Alec R. Chapmanb,d, Jun Zhaoc, Liya Xua, Tongtong Anc,QiMaa, Yuyan Wangc, Meina Wuc, Yu Sune, Shuhang Wangc, Zhenxiang Lic, Xiaodan Yangc, Jun Yongb, Xiao-Dong Sua, Youyong Luf, Fan Baia,3, X. Sunney Xiea,b,3, and Jie Wangc,3 aBiodynamic Optical Imaging Center, School of Life Sciences, Peking University, Beijing 100871, China; bDepartment of Chemistry and Chemical Biology, dProgram in Biophysics, Harvard University, Cambridge, MA 02138; and Departments of cThoracic Medical Oncology and ePathology, fLaboratory of Molecular Oncology, Key Laboratory of Carcinogenesis and Translational Research (Ministry of Education), Peking University Cancer Hospital and Institute, Beijing 100142, China Contributed by X. Sunney Xie, November 5, 2013 (sent for review September 28, 2013) Circulating tumor cells (CTCs) enter peripheral blood from primary 11 patients (SI Appendix, Table S1) with lung cancer, the leading tumors and seed metastases. The genome sequencing of CTCs cause of worldwide cancer-related deaths. CTCs were captured could offer noninvasive prognosis or even diagnosis, but has been with the CellSearch platform using antibodies enrichment after hampered by low single-cell genome coverage of scarce CTCs. fixation, further isolated with 94% specificity (Materials and Here, we report the use of the recently developed multiple anneal- Methods), and then subjected to WGA using MALBAC before ing and looping-based amplification cycles for whole-genome am- next-generation sequencing. -

Expression Pattern of Olfactory Receptor Genes in Human Cumulus Cells As an Indicator for Competent Oocyte Selection
Turkish Journal of Biology Turk J Biol (2020) 44: 371-380 http://journals.tubitak.gov.tr/biology/ © TÜBİTAK Research Article doi:10.3906/biy-2003-79 Expression pattern of olfactory receptor genes in human cumulus cells as an indicator for competent oocyte selection 1 2 1,3 4 5 Neda DAEI-FARSHBAF , Reza AFLATOONIAN , Fatemeh-Sadat AMJADI , Sara TALEAHMAD , Mahnaz ASHRAFI , 3,1, Mehrdad BAKHTIYARI * 1 Department of Anatomy, Faculty of Medicine, Iran University of Medical Sciences, Tehran, Iran 2 Department of Endocrinology and Female Infertility, Reproductive Biomedicine Research Center, Royan Institute for Reproductive Biomedicine, Academic Center for Education, Culture and Research, Tehran, Iran 3 Cellular and Molecular Research Center, Faculty of Medicine, Iran University of Medical Sciences, Tehran, Iran 4 Department of Molecular Systems Biology, Cell Science Research Center, Royan Institute for Stem Cell Biology and Technology (RI-SCBT), Academic Center for Education, Culture and Research, Tehran, Iran 5 Department of Obstetrics and Gynecology, Faculty of Medicine, Iran University of Medical Sciences, Tehran, Iran Received: 26.03.2020 Accepted/Published Online: 09.09.2020 Final Version: 14.12.2020 Abstract: Odorant or olfactory receptors are mainly localized in the olfactory epithelium for the perception of different odors. Interestingly, many ectopic olfactory receptors with low expression levels have recently been found in nonolfactory tissues to involve in local functions. Therefore, we investigated the probable role of the olfactory signaling pathway in the surrounding microenvironment of oocyte. This study included 22 women in intracytoplasmic sperm injection cycle. The expression of olfactory target molecules in cumulus cells surrounding the growing and mature oocytes was evaluated by Western blotting and real-time polymerase chain reaction.