Gene Section Review
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplemental Information to Mammadova-Bach Et Al., “Laminin Α1 Orchestrates VEGFA Functions in the Ecosystem of Colorectal Carcinogenesis”
Supplemental information to Mammadova-Bach et al., “Laminin α1 orchestrates VEGFA functions in the ecosystem of colorectal carcinogenesis” Supplemental material and methods Cloning of the villin-LMα1 vector The plasmid pBS-villin-promoter containing the 3.5 Kb of the murine villin promoter, the first non coding exon, 5.5 kb of the first intron and 15 nucleotides of the second villin exon, was generated by S. Robine (Institut Curie, Paris, France). The EcoRI site in the multi cloning site was destroyed by fill in ligation with T4 polymerase according to the manufacturer`s instructions (New England Biolabs, Ozyme, Saint Quentin en Yvelines, France). Site directed mutagenesis (GeneEditor in vitro Site-Directed Mutagenesis system, Promega, Charbonnières-les-Bains, France) was then used to introduce a BsiWI site before the start codon of the villin coding sequence using the 5’ phosphorylated primer: 5’CCTTCTCCTCTAGGCTCGCGTACGATGACGTCGGACTTGCGG3’. A double strand annealed oligonucleotide, 5’GGCCGGACGCGTGAATTCGTCGACGC3’ and 5’GGCCGCGTCGACGAATTCACGC GTCC3’ containing restriction site for MluI, EcoRI and SalI were inserted in the NotI site (present in the multi cloning site), generating the plasmid pBS-villin-promoter-MES. The SV40 polyA region of the pEGFP plasmid (Clontech, Ozyme, Saint Quentin Yvelines, France) was amplified by PCR using primers 5’GGCGCCTCTAGATCATAATCAGCCATA3’ and 5’GGCGCCCTTAAGATACATTGATGAGTT3’ before subcloning into the pGEMTeasy vector (Promega, Charbonnières-les-Bains, France). After EcoRI digestion, the SV40 polyA fragment was purified with the NucleoSpin Extract II kit (Machery-Nagel, Hoerdt, France) and then subcloned into the EcoRI site of the plasmid pBS-villin-promoter-MES. Site directed mutagenesis was used to introduce a BsiWI site (5’ phosphorylated AGCGCAGGGAGCGGCGGCCGTACGATGCGCGGCAGCGGCACG3’) before the initiation codon and a MluI site (5’ phosphorylated 1 CCCGGGCCTGAGCCCTAAACGCGTGCCAGCCTCTGCCCTTGG3’) after the stop codon in the full length cDNA coding for the mouse LMα1 in the pCIS vector (kindly provided by P. -

Gene Symbol Gene Description ACVR1B Activin a Receptor, Type IB
Table S1. Kinase clones included in human kinase cDNA library for yeast two-hybrid screening Gene Symbol Gene Description ACVR1B activin A receptor, type IB ADCK2 aarF domain containing kinase 2 ADCK4 aarF domain containing kinase 4 AGK multiple substrate lipid kinase;MULK AK1 adenylate kinase 1 AK3 adenylate kinase 3 like 1 AK3L1 adenylate kinase 3 ALDH18A1 aldehyde dehydrogenase 18 family, member A1;ALDH18A1 ALK anaplastic lymphoma kinase (Ki-1) ALPK1 alpha-kinase 1 ALPK2 alpha-kinase 2 AMHR2 anti-Mullerian hormone receptor, type II ARAF v-raf murine sarcoma 3611 viral oncogene homolog 1 ARSG arylsulfatase G;ARSG AURKB aurora kinase B AURKC aurora kinase C BCKDK branched chain alpha-ketoacid dehydrogenase kinase BMPR1A bone morphogenetic protein receptor, type IA BMPR2 bone morphogenetic protein receptor, type II (serine/threonine kinase) BRAF v-raf murine sarcoma viral oncogene homolog B1 BRD3 bromodomain containing 3 BRD4 bromodomain containing 4 BTK Bruton agammaglobulinemia tyrosine kinase BUB1 BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) BUB1B BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) C9orf98 chromosome 9 open reading frame 98;C9orf98 CABC1 chaperone, ABC1 activity of bc1 complex like (S. pombe) CALM1 calmodulin 1 (phosphorylase kinase, delta) CALM2 calmodulin 2 (phosphorylase kinase, delta) CALM3 calmodulin 3 (phosphorylase kinase, delta) CAMK1 calcium/calmodulin-dependent protein kinase I CAMK2A calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha CAMK2B calcium/calmodulin-dependent -

Methods in and Applications of the Sequencing of Short Non-Coding Rnas" (2013)
University of Pennsylvania ScholarlyCommons Publicly Accessible Penn Dissertations 2013 Methods in and Applications of the Sequencing of Short Non- Coding RNAs Paul Ryvkin University of Pennsylvania, [email protected] Follow this and additional works at: https://repository.upenn.edu/edissertations Part of the Bioinformatics Commons, Genetics Commons, and the Molecular Biology Commons Recommended Citation Ryvkin, Paul, "Methods in and Applications of the Sequencing of Short Non-Coding RNAs" (2013). Publicly Accessible Penn Dissertations. 922. https://repository.upenn.edu/edissertations/922 This paper is posted at ScholarlyCommons. https://repository.upenn.edu/edissertations/922 For more information, please contact [email protected]. Methods in and Applications of the Sequencing of Short Non-Coding RNAs Abstract Short non-coding RNAs are important for all domains of life. With the advent of modern molecular biology their applicability to medicine has become apparent in settings ranging from diagonistic biomarkers to therapeutics and fields angingr from oncology to neurology. In addition, a critical, recent technological development is high-throughput sequencing of nucleic acids. The convergence of modern biotechnology with developments in RNA biology presents opportunities in both basic research and medical settings. Here I present two novel methods for leveraging high-throughput sequencing in the study of short non- coding RNAs, as well as a study in which they are applied to Alzheimer's Disease (AD). The computational methods presented here include High-throughput Annotation of Modified Ribonucleotides (HAMR), which enables researchers to detect post-transcriptional covalent modifications ot RNAs in a high-throughput manner. In addition, I describe Classification of RNAs by Analysis of Length (CoRAL), a computational method that allows researchers to characterize the pathways responsible for short non-coding RNA biogenesis. -

14-3-3 Proteins Inactivate DAPK2 by Promoting Its Dimerization And
ARTICLE https://doi.org/10.1038/s42003-021-02518-y OPEN 14-3-3 proteins inactivate DAPK2 by promoting its dimerization and protecting key regulatory phosphosites ✉ ✉ Matej Horvath 1,2, Olivia Petrvalska1,2, Petr Herman 3, Veronika Obsilova 2 & Tomas Obsil 1,2 Death-associated protein kinase 2 (DAPK2) is a CaM-regulated Ser/Thr protein kinase, involved in apoptosis, autophagy, granulocyte differentiation and motility regulation, whose activity is controlled by autoinhibition, autophosphorylation, dimerization and interaction with scaffolding proteins 14-3-3. However, the structural basis of 14-3-3-mediated DAPK2 reg- ulation remains unclear. Here, we structurally and biochemically characterize the full-length 1234567890():,; human DAPK2:14-3-3 complex by combining several biophysical techniques. The results from our X-ray crystallographic analysis revealed that Thr369 phosphorylation at the DAPK2 C terminus creates a high-affinity canonical mode III 14-3-3-binding motif, further enhanced by the diterpene glycoside Fusicoccin A. Moreover, concentration-dependent DAPK2 dimerization is disrupted by Ca2+/CaM binding and stabilized by 14-3-3 binding in solution, thereby protecting the DAPK2 inhibitory autophosphorylation site Ser318 against depho- sphorylation and preventing Ca2+/CaM binding. Overall, our findings provide mechanistic insights into 14-3-3-mediated DAPK2 inhibition and highlight the potential of the DAPK2:14- 3-3 complex as a target for anti‐inflammatory therapies. 1 Department of Physical and Macromolecular Chemistry, Faculty of Science, Charles University, Prague, Czech Republic. 2 Department of Structural Biology of Signaling Proteins, Division BIOCEV, Institute of Physiology of the Czech Academy of Sciences, Vestec, Czech Republic. 3 Institute of Physics, Faculty of ✉ Mathematics and Physics, Charles University, Prague, Czech Republic. -

Human Cumulus Cells in Long-Term in Vitro Culture Reflect Differential
cells Article Human Cumulus Cells in Long-Term In Vitro Culture Reflect Differential Expression Profile of Genes Responsible for Planned Cell Death and Aging—A Study of New Molecular Markers Bła˙zejChermuła 1, Wiesława Kranc 2 , Karol Jopek 3, Joanna Budna-Tukan 3 , Greg Hutchings 2,4, Claudia Dompe 3,4, Lisa Moncrieff 3,4 , Krzysztof Janowicz 2,4, Małgorzata Józkowiak 5, Michal Jeseta 6 , Jim Petitte 7 , Paul Mozdziak 8 , Leszek Pawelczyk 1, Robert Z. Spaczy ´nski 1 and Bartosz Kempisty 2,3,6,9,* 1 Division of Infertility and Reproductive Endocrinology, Department of Gynecology, Obstetrics and Gynecological Oncology, Poznan University of Medical Sciences, 33 Polna St., 60-535 Poznan, Poland; [email protected] (B.C.); [email protected] (L.P.); [email protected] (R.Z.S.) 2 Department of Anatomy, Poznan University of Medical Sciences, 6 Swiecickiego St., 60-781 Poznan, Poland; [email protected] (W.K.); [email protected] (G.H.); [email protected] (K.J.) 3 Department of Histology and Embryology, Poznan University of Medical Sciences, 6 Swiecickiego St., 60-781 Poznan, Poland; [email protected] (K.J.); [email protected] (J.B.-T.); [email protected] (C.D.); l.moncrieff[email protected] (L.M.) 4 The School of Medicine, Medical Sciences and Nutrition, University of Aberdeen, Aberdeen AB25 2ZD, UK 5 Department of Toxicology, Poznan University of Medical Sciences, 30 Dojazd St., 60-631 Poznan, Poland; [email protected] 6 Department of Obstetrics and Gynecology, University Hospital and -

Cerebral Cavernous Malformations: from Genes to Proteins to Disease
See the corresponding editorial in this issue, pp 119–121. J Neurosurg 116:122–132, 2012 Cerebral cavernous malformations: from genes to proteins to disease Clinical article DANIEL D. CAVALCANTI, M.D., M. YASHAR S. KALANI, M.D., PH.D., NIKOLAY L. MARTIROSYAN, M.D., JUSTIN EALES, B.S., ROBERT F. SPETZLER, M.D., AND MARK C. PREUL, M.D. Division of Neurological Surgery, Barrow Neurological Institute, St. Joseph’s Hospital and Medical Center, Phoenix, Arizona Over the past half century molecular biology has led to great advances in our understanding of angio- and vas- culogenesis and in the treatment of malformations resulting from these processes gone awry. Given their sporadic and familial distribution, their developmental and pathological link to capillary telangiectasias, and their observed chromosomal abnormalities, cerebral cavernous malformations (CCMs) are regarded as akin to cancerous growths. Although the exact pathological mechanisms involved in the formation of CCMs are still not well understood, the identification of 3 genetic loci has begun to shed light on key developmental pathways involved in CCM pathogen- esis. Cavernous malformations can occur sporadically or in an autosomal dominant fashion. Familial forms of CCMs have been attributed to mutations at 3 different loci implicated in regulating important processes such as proliferation and differentiation of angiogenic precursors and members of the apoptotic machinery. These processes are important for the generation, maintenance, and pruning of every vessel in the body. In this review the authors highlight the lat- est discoveries pertaining to the molecular genetics of CCMs, highlighting potential new therapeutic targets for the treatment of these lesions. -

Novel Functions of Death-Associated Protein Kinases Through Mitogen-Activated Protein Kinase-Related Signals
International Journal of Molecular Sciences Article Novel Functions of Death-Associated Protein Kinases through Mitogen-Activated Protein Kinase-Related Signals Mohamed Elbadawy 1,2,† , Tatsuya Usui 1,*,†, Hideyuki Yamawaki 3 and Kazuaki Sasaki 1 1 Laboratory of Veterinary Pharmacology, Department of Veterinary Medicine, Faculty of Agriculture, Tokyo University of Agriculture and Technology, 3-5-8 Saiwai-cho, Fuchu, Tokyo 183-8509, Japan; [email protected] (M.E.); [email protected] (K.S.) 2 Department of Pharmacology, Faculty of Veterinary Medicine, Benha University, Moshtohor, Elqaliobiya, Toukh 13736, Egypt 3 Laboratory of Veterinary Pharmacology, School of Veterinary Medicine, Kitasato University, Towada, Aomori 034-8628, Japan; [email protected] * Correspondence: [email protected]; Tel./Fax: +81-42-367-5769 † These authors contributed equally to this work. Received: 13 September 2018; Accepted: 1 October 2018; Published: 4 October 2018 Abstract: Death associated protein kinase (DAPK) is a calcium/calmodulin-regulated serine/threonine kinase; its main function is to regulate cell death. DAPK family proteins consist of DAPK1, DAPK2, DAPK3, DAPK-related apoptosis-inducing protein kinases (DRAK)-1 and DRAK-2. In this review, we discuss the roles and regulatory mechanisms of DAPK family members and their relevance to diseases. Furthermore, a special focus is given to several reports describing cross-talks between DAPKs and mitogen-activated protein kinases (MAPK) family members in various pathologies. We also discuss small molecule inhibitors of DAPKs and their potential as therapeutic targets against human diseases. Keywords: MAPK; DAPK; ERK; p38; JNK 1. Introduction: DAPKs, MAPKs Death-associated protein kinase (DAPK) family proteins are closely related, Ca2+/calmodulin (CaM)-regulated serine/threonine kinases, whose members not only possess significant homology in their catalytic domains but also share cell death-associated functions [1,2]. -

Anti-DAPK2 Antibody (ARG54388)
Product datasheet [email protected] ARG54388 Package: 50 μg anti-DAPK2 antibody Store at: -20°C Summary Product Description Rabbit Polyclonal antibody recognizes DAPK2 Tested Reactivity Hu, Ms, Rat Tested Application IHC, WB Specificity This antibody recognizes human, mouse, and rat DAPK2 (approx. 42kDa) and does not cross-react with DAPK. Host Rabbit Clonality Polyclonal Isotype IgG Target Name DAPK2 Antigen Species Human Immunogen Peptide corresponding to aa 356-370 of human DAPK2 (accession no. BAA88063). This sequence is identical to that of mouse. Conjugation Un-conjugated Alternate Names DRP1; DAP-kinase-related protein 1; DRP-1; DAP kinase 2; EC 2.7.11.1; Death-associated protein kinase 2 Application Instructions Application table Application Dilution IHC Assay-dependent WB Assay-dependent Application Note * The dilutions indicate recommended starting dilutions and the optimal dilutions or concentrations should be determined by the scientist. Positive Control A431, Mouse spleen and Rat kidney Calculated Mw 43 kDa Properties Form Liquid Purification Immunoaffinity chroma-tography Buffer PBS (pH 7.4) and 0.02% Sodium azide Preservative 0.02% Sodium azide Storage instruction For continuous use, store undiluted antibody at 2-8°C for up to a week. For long-term storage, aliquot and store at -20°C or below. Storage in frost free freezers is not recommended. Avoid repeated freeze/thaw cycles. Suggest spin the vial prior to opening. The antibody solution should be gently mixed www.arigobio.com 1/3 before use. Note For laboratory research only, not for drug, diagnostic or other use. Bioinformation Database links GeneID: 13143 Mouse GeneID: 23604 Human Swiss-port # Q8VDF3 Mouse Swiss-port # Q9UIK4 Human Gene Symbol DAPK2 Gene Full Name death-associated protein kinase 2 Background Certain serine/threonine protein kinases, such as RIP and DAP kinase, are mediators of apoptosis. -

Supplementary Information
Supplementary Information Table S1. Gene Ontology analysis of affected biological processes/pathways/themes in H1 hESCs based on sets of statistically significant differentially expressed genes. Exposures Overrepresented Categories (Upregulation) EASE Score Negative regulation of cell differentiation 0.0049 Lipid biosynthetic process 0.015 Negative regulation of cell proliferation 0.02 5 cGy, 2 h Transcription factor binding 0.037 Regulation of apoptosis 0.038 Positive regulation of anti-apoptosis 0.048 P53 signaling pathway 4.2 × 10−10 Positive regulation of apoptosis 7.5 × 10−8 Response to DNA damage stimulus 1.5 × 10−6 Cellular response to stress 9.6 × 10−6 1 Gy, 2 h Negative regulation of cell proliferation 9.8 × 10−6 Cell cycle arrest 7.0 × 10−4 Negative regulation of cell differentiation 1.5 × 10−3 Regulation of protein kinase activity 0.011 I-kappaB kinase/NF-kappaB cascade 0.025 Metallothionein superfamily 9.9 × 10−18 Induction of apoptosis 8.8 × 10−5 DNA damage response 2.6 × 10−4 1 Gy, 16 h Positive regulation of anti-apoptosis 0.001 Positive regulation of cell death 0.005 Cellular response to stress 0.012 Exposures Overrepresented Categories (Downregulation) EASE Score Alternative splicing 0.016 1 Gy, 2 h Chromatin organization 0.020 Chromatin assembly 2.0 × 10−6 Cholesterol biosynthesis 5.1 × 10−5 Macromolecular complex assembly 9.3 × 10−4 1 Gy, 16 h PPAR signaling pathway 0.007 Hemopoietic organ development 0.033 Immune system development 0.040 S2 Table S2. Gene Ontology analysis of affected biological processes/pathways/themes in H7 based on sets of statistically significant differentially expressed genes. -

Context-Selective Death of Acute Myeloid Leukemia Cells Triggered by the Novel Hybrid Retinoid-HDAC Inhibitor MC2392
Published OnlineFirst February 24, 2014; DOI: 10.1158/0008-5472.CAN-13-2568 Cancer Therapeutics, Targets, and Chemical Biology Research Context-Selective Death of Acute Myeloid Leukemia Cells Triggered by the Novel Hybrid Retinoid-HDAC Inhibitor MC2392 Floriana De Bellis1,4, Vincenzo Carafa1, Mariarosaria Conte1, Dante Rotili3, Francesca Petraglia1, Filomena Matarese4, Kees-Jan Francoijs¸ 4, Julien Ablain6, Sergio Valente3,Remy Castellano5, Armelle Goubard5, Yves Collette5, Amit Mandoli4, Joost H.A. Martens4, Hugues de The6, Angela Nebbioso1, Antonello Mai3, Hendrik G. Stunnenberg4, and Lucia Altucci1,2 Abstract HDAC inhibitors (HDACi) are widely used in the clinic to sensitize tumorigenic cells for treatment with other anticancer compounds. The major drawback of HDACi is the broad inhibition of the plethora of HDAC-containing complexes. In acute promyelocytic leukemia (APL), repression by the PML-RARa oncofusion protein is mediated by an HDAC-containing complex that can be dissociated by pharmacologic doses of all trans retinoic acid (ATRA) inducing differentiation and cell death at the expense of side effects and recurrence. We hypothesized that the context-specific close physical proximity of a retinoid and HDACi-binding protein in the repressive PML-RARa- HDAC complex may permit selective targeting by a hybrid molecule of ATRA with a 2-aminoanilide tail of the HDAC inhibitor MS-275, yielding MC2392. We show that MC2392 elicits weak ATRA and essentially no HDACi activity in vitro or in vivo. Genome-wide epigenetic analyses revealed that in NB4 cells expressing PML-RARa, MC2392 induces changes in H3 acetylation at a small subset of PML-RARa–binding sites. RNA-seq reveals that MC2392 alters expression of a number of stress-responsive and apoptotic genes. -
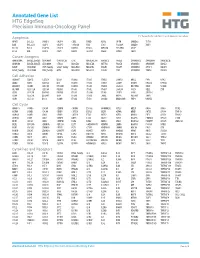
Annotated Gene List HTG Edgeseq Precision Immuno-Oncology Panel
Annotated Gene List HTG EdgeSeq Precision Immuno-Oncology Panel For Research Use Only. Not for use in diagnostic procedures. Apoptosis APAF1 BCL2L1 CARD11 CASP4 CD5L FADD KSR2 OPTN SAMD12 TCF19 BAX BCL2L11 CASP1 CASP5 CORO1A FAS LRG1 PLA2G6 SAMD9 XAF1 BCL10 BCL6 CASP10 CASP8 DAPK2 FASLG MECOM PYCARD SPOP BCL2 BID CASP3 CAV1 DAPL1 GLIPR1 MELK RIPK2 TBK1 Cancer Antigens ANKRD30A BAGE2_BAGE3 CEACAM6 CTAG1A_1B LIPE MAGEA3_A6 MAGEC2 PAGE3 SPANXACD SPANXN4 XAGE1B_1E ARMCX6 BAGE4_BAGE5 CEACAM8 CTAG2 MAGEA1 MAGEA4 MTFR2 PAGE4 SPANXB1 SPANXN5 XAGE2 BAGE CEACAM1 CT45_family GAGE_family MAGEA10 MAGEB2 PAGE1 PAGE5 SPANXN1 SYCP1 XAGE3 BAGE_family CEACAM5 CT47_family HPN MAGEA12 MAGEC1 PAGE2 PBK SPANXN3 TEX14 XAGE5 Cell Adhesion ADAM17 CDH15 CLEC5A DSG3 ICAM2 ITGA5 ITGB2 LAMC3 MBL2 PVR UPK2 ADD2 CDH5 CLEC6A DST ICAM3 ITGA6 ITGB3 LAMP1 MTDH RRAS2 UPK3A ADGRE5 CLDN3 CLEC7A EPCAM ICAM4 ITGAE ITGB4 LGALS1 NECTIN2 SELE VCAM1 ALCAM CLEC12A CLEC9A FBLN1 ITGA1 ITGAL ITGB7 LGALS3 OCLN SELL ZYX CD63 CLEC2B DIAPH3 FXYD5 ITGA2 ITGAM ITLN2 LYVE1 OLR1 SELPLG CD99 CLEC4A DLGAP5 IBSP ITGA3 ITGAX JAML M6PR PECAM1 THY1 CDH1 CLEC4C DSC3 ICAM1 ITGA4 ITGB1 L1CAM MADCAM1 PKP1 UNC5D Cell Cycle ANAPC1 CCND3 CDCA5 CENPH CNNM1 ESCO2 HORMAD2 KIF2C MELK ORC6 SKA3 TPX2 ASPM CCNE1 CDCA8 CENPI CNTLN ESPL1 IKZF1 KIF4A MND1 PATZ1 SP100 TRIP13 AURKA CCNE2 CDK1 CENPL CNTLN ETS1 IKZF2 KIF5C MYBL2 PIF1 SP110 TROAP AURKB CCNF CDK4 CENPU DBF4 ETS2 IKZF3 KIFC1 NCAPG PIMREG SPC24 TUBB BEX1 CDC20 CDK6 CENPW E2F2 EZH2 IKZF4 KNL1 NCAPG2 PKMYT1 SPC25 ZWILCH BEX2 CDC25A CDKN1A CEP250 E2F7 GADD45GIP1 -

PRODUCTS and SERVICES Target List
PRODUCTS AND SERVICES Target list Kinase Products P.1-11 Kinase Products Biochemical Assays P.12 "QuickScout Screening Assist™ Kits" Kinase Protein Assay Kits P.13 "QuickScout Custom Profiling & Panel Profiling Series" Targets P.14 "QuickScout Custom Profiling Series" Preincubation Targets Cell-Based Assays P.15 NanoBRET™ TE Intracellular Kinase Cell-Based Assay Service Targets P.16 Tyrosine Kinase Ba/F3 Cell-Based Assay Service Targets P.17 Kinase HEK293 Cell-Based Assay Service ~ClariCELL™ ~ Targets P.18 Detection of Protein-Protein Interactions ~ProbeX™~ Stable Cell Lines Crystallization Services P.19 FastLane™ Structures ~Premium~ P.20-21 FastLane™ Structures ~Standard~ Kinase Products For details of products, please see "PRODUCTS AND SERVICES" on page 1~3. Tyrosine Kinases Note: Please contact us for availability or further information. Information may be changed without notice. Expression Protein Kinase Tag Carna Product Name Catalog No. Construct Sequence Accession Number Tag Location System HIS ABL(ABL1) 08-001 Full-length 2-1130 NP_005148.2 N-terminal His Insect (sf21) ABL(ABL1) BTN BTN-ABL(ABL1) 08-401-20N Full-length 2-1130 NP_005148.2 N-terminal DYKDDDDK Insect (sf21) ABL(ABL1) [E255K] HIS ABL(ABL1)[E255K] 08-094 Full-length 2-1130 NP_005148.2 N-terminal His Insect (sf21) HIS ABL(ABL1)[T315I] 08-093 Full-length 2-1130 NP_005148.2 N-terminal His Insect (sf21) ABL(ABL1) [T315I] BTN BTN-ABL(ABL1)[T315I] 08-493-20N Full-length 2-1130 NP_005148.2 N-terminal DYKDDDDK Insect (sf21) ACK(TNK2) GST ACK(TNK2) 08-196 Catalytic domain