Multiple Pathways to Homothallism in Closely Related Yeast Lineages in the Basidiomycota 2 3 Alexandra Cabrita1, Márcia David-Palma1*, Patrícia H
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Evolution of Genetic Systems in Filamentous Ascomycetes
Evolution of Genetic Systems in Filamentous Ascomycetes Evolutie van genetische systemen in hyphenvormende zakjeszwammen 0000 0513 3836 Promotor: dr. R.F. Hoekstra hoogleraar in de populatie- en kwantitatieve genetica fjtfoiißi f ßin Maarten J. Nauta Evolution of Genetic Systems in Filamentous Ascomycetes Proefschrift ter verkrijging van de graad van doctor in de landbouw- en milieuwetenschappen op gezag van de rector magnificus, dr. C.M. Karssen, in het openbaar te verdedigen op woensdag 12januar i 1994 des namiddags te vier uur in de Aula van de Landbouwuniversiteit te Wageningen. 15 0 S(p^ZJ> These investigations were supported by the Netherlands Organization for Scientific Research (N.W.O.). BibUt/FHEEK LAMDbOirWUNIVERSITEJi. WAGE NINGE N CIP-GEGEVENS KONINKLIJKE BIBLIOTHEEK, DEN HAAG Nauta, Maarten J. Evolution of genetic systems in filamentous ascomycetes / Maarten J. Nauta. - [ S.l. : s.n.]. -111 . Thesis Wageningen. - With ref. - With summary in Dutch. ISBN 90-5485-199-6 Subject headings: population genetics / ascomycetes. omslagontwerp: Ernst van Cleef foto omslag: Barrages tussen verschillende stammen van Podospora anserina als gevolg van vegetatieve incompatibiliteit. (met dank aan Inge Haspels) aan mijn ouders Voorwoord Dit proefschrift is het resultaat van vier jaar onderzoek, verricht bij de vakgroep Erfelijkheidsleer van de Landbouwuniversiteit in Wageningen. In zekere zin valt zo'n proefschrift te vergelijken met een levend wezen. Uit de genetica is bekend dat de verschijningsvorm van elk levend wezen tot stand komt door een combinatie van erfelijke aanleg en invloeden uit de omgeving. Voor een proefschrift geldt eigenlijk hetzelfde: Zowel het werk van de auteur, als de bijdragen van zijn omgeving zijn onontbeerlijk om tot een verschijningsvorm te komen. -

Algal Sex Determination and the Evolution of Anisogamy James Umen, Susana Coelho
Algal Sex Determination and the Evolution of Anisogamy James Umen, Susana Coelho To cite this version: James Umen, Susana Coelho. Algal Sex Determination and the Evolution of Anisogamy. Annual Review of Microbiology, Annual Reviews, 2019, 73 (1), 10.1146/annurev-micro-020518-120011. hal- 02187088 HAL Id: hal-02187088 https://hal.sorbonne-universite.fr/hal-02187088 Submitted on 17 Jul 2019 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Annu. Rev. Microbiol. 2019. 73:X–X https://doi.org/10.1146/annurev-micro-020518-120011 Copyright © 2019 by Annual Reviews. All rights reserved Umen • Coelho www.annualreviews.org • Algal Sexes and Mating Systems Algal Sex Determination and the Evolution of Anisogamy James Umen1 and Susana Coelho2 1Donald Danforth Plant Science Center, St. Louis, Missouri 63132, USA; email: [email protected] 2Sorbonne Université, UPMC Université Paris 06, CNRS, Algal Genetics Group, UMR 8227, Integrative Biology of Marine Models, Station Biologique de Roscoff, CS 90074, F-29688, Roscoff, France [**AU: Please write the entire affiliation in French or write it all in English, rather than a combination of English and French**] ; email: [email protected] Abstract Algae are photosynthetic eukaryotes whose taxonomic breadth covers a range of life histories, degrees of cellular and developmental complexity, and diverse patterns of sexual reproduction. -

Molecular Mechanisms of Sexual Reproduction in the Order Cystofilobasidiales
Alexandra Sofia Rodrigues Cabrita Licenciada em Biologia Celular e Molecular Molecular Mechanisms of Sexual Reproduction in the Order Cystofilobasidiales Dissertação para obtenção do Grau de Mestre em Genética Molecular e Biomedicina Orientadora: Professora Doutora Paula Gonçalves, Universidade Nova de Lisboa Co-orientador: Doutor Marco Coelho, Universidade Nova de Lisboa Júri: Presidente: Professora Doutora Margarida Casal Ribeiro Castro Caldas Braga Arguente: Doutora Maria Teresa Proença Mendes Maia Vogal: Professora Doutora Paula Maria Theriaga Mendes Bernardo Gonçalves Novembro, 2018 Alexandra Sofia Rodrigues Cabrita Licenciada em Biologia Celular e Molecular Molecular Mechanisms of Sexual Reproduction in the Order Cystofilobasidiales Dissertação para obtenção do Grau de Mestre em Genética Molecular e Biomedicina Orientadora: Professora Doutora Paula Gonçalves, Universidade Nova de Lisboa Co-orientador: Doutor Marco Coelho, Universidade Nova de Lisboa Júri: Presidente: Professora Doutora Margarida Casal Ribeiro Castro Caldas Braga Arguente: Doutora Maria Teresa Proença Mendes Maia Vogal: Professora Doutora Paula Maria Theriaga Mendes Bernardo Gonçalves Novembro, 2018 Molecular Mechanisms of Sexual Reproduction in the Order Cystofilobasidiales Copyright Alexandra Sofia Rodrigues Cabrita, FCT/UNL, UNL A Faculdade de Ciências e Tecnologia e a Universidade Nova de Lisboa têm o direito, perpétuo e sem limites geográficos, de arquivar e publicar esta dissertação através de exemplares impressos reproduzidos em papel ou de forma digital, ou por qualquer outro meio conhecido ou que venha a ser inventado, e de a divulgar através de repositórios científicos e de admitir a sua cópia e distribuição com objectivos educacionais ou de investigação, não comerciais, desde que seja dado crédito ao autor e editor. ii Acknowledgments First, I would like to thank my supervisor, Professor Paula Gonçalves, for having hosted me in the Yeast Genomics lab and for all the help and support throughout this year. -

Sex in the Extremes: Lichen-Forming Fungi
Mycologist, Volume 19, Part 2 May 2005. ©Cambridge University Press Printed in the United Kingdom. DOI: 10.1017/S0269915XO5002016 Sex in the extremes: lichen-forming fungi FABIAN A. SEYMOUR, PETER D. CRITTENDEN & PAUL S. DYER* School of Biology, University of Nottingham, University Park, Nottingham, NG7 2RD, UK. Tel. +44 (0) 115 9513203, Fax +44 (0) 115 9513251 E-mail: [email protected]; [email protected] *Corresponding Author Lichens are characteristically found in environments subject to extremes of temperature, desiccation and low nutrient status. Despite this sexual structures are often formed in abundance. The underlying mechanisms of sex in lichen-forming fungi are discussed, together with possible ecological reasons for the persistence of sexuality. Special features of lichen sex are highlighted including sex at the limits of life on earth in Antarctica, re-licheniza- tion following sex and dispersal, and the perennial nature of lichen fruiting bodies. Keywords: lichen, fungi, sex, breeding system, (98%) belonging to the Ascomycotina (Kirk et al., symbiosis, extreme environments, Antarctica 2001). They display a variety of morphologies, from flattened crust (crustose) or leafy (foliose) forms to Lichens - living together in a long-term relation- shrubby or pendulous fruticose types (Honegger, 2001) ship (Figs 3, 4, 7, 8). Lichens are seen as a textbook example of a successful Life in extreme environments mutualistic symbiosis. They consist of at least two A key characteristic of lichens is that they have a organisms: a fungus (the ‘mycobiont’), and an remarkable ability to tolerate extreme environmental intimately associated photosynthetic partner (the conditions and sustain growth despite frequent cycles ‘photobiont’). -

Udeniomyces Kanasensis Sp. Nov., a Ballistoconidium-Forming Yeast Species in the Cystofilobasidiales
Antonie van Leeuwenhoek (2012) 102:45–51 DOI 10.1007/s10482-012-9711-5 ORIGINAL PAPER Udeniomyces kanasensis sp. nov., a ballistoconidium-forming yeast species in the Cystofilobasidiales Pei-Jie Han • Jun-Zhi Qiu • Qi-Ming Wang • Feng-Yan Bai Received: 10 November 2011 / Accepted: 4 February 2012 / Published online: 22 February 2012 Ó Springer Science+Business Media B.V. 2012 Abstract In a survey of ballistoconidium-forming Abbreviation yeast diversity in the phyllosphere, five strains from ITS Internal transcribed spacer wilting plant leaves collected from Kanas Nature Reserve in Xinjiang province, China were selected based on morphological comparison. These strains Introduction formed pinkish-white colonies and large bilaterally symmetrical ballistoconidia. Molecular phylogenetic The genus Udeniomyces was proposed for threes analyses based on the 26S rRNA D1/D2 domain and species, Udeniomyces megalosporus, Udeniomyces pu- ITS region sequences showed that these strains niceus and Udeniomyces pyricola (Nakase and Tak- belonged to the Udeniomyces clade in the Cystofi- ematsu 1992), which were classified previously in the lobasidiales. They differ from the described Udeni- ‘pyricola group’ of the genus Bullera (Nakase 1987)and omyces species significantly in the rRNA sequences as characterized by forming large bilaterally symmetrical well as physiological criteria. Therefore, a new species ballistoconidia and pinkish-white to pale pink colonies Udeniomyces kanasensis sp. nov. (type strain XJ (Nakase 1989). Niwata et al. (2002)describedUdeni- 6E2T=CGMCC 2.02627 T=CBS 12488 T) is proposed omyces pannonicus on the basis of its morphological to accommodate these strains. The MycoBank number and chemotaxonomic characteristics. However, this of the new species is MB 563659. -
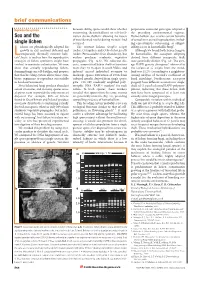
Brief Comms Layout 6/4Mx
brief communications Reproductive systems between sibling spores would show whether perpetuates successful genotypes adapted to outcrossing (heterothallism) or self-fertil- the prevailing environmental regimes. Sex and the ization (homothallism: allowing the fusion Homothallism also retains certain benefits of two identical nuclei during meiosis) had of sexual over asexual reproduction, includ- single lichen occurred. ing opportunistic outcrossing, as obligate ichens are physiologically adapted for The crustose lichens Graphis scripta selfing is rare in homothallic fungi8. growth in dry, nutrient-deficient and (order: Ostropales) and Ochrolechia parella Although we found both lichen fungi to Ltemporarily thermally extreme habi- (order: Pertusariales) fruit abundantly, but be homothallic, the ascospore offspring tats1, but it is unclear how the reproductive neither produces symbiotic vegetative derived from different conspecific thalli strategies of lichen symbionts might have propagules (Fig. 1a,b). We collected dis- were genetically distinct (Fig. 1d). The aver- evolved to maximize colonization. We now crete, symmetrical lichen thalli at locations age RAPD genetic divergence9 observed in show that sexually reproducing lichen- more than 10 m apart in south Wales, and five isolates of G. scripta from one wood- forming fungi can self-fertilize, and propose induced excised individual ascomata to land was 15.2% (according to a neighbour- that this breeding system allows these sym- discharge spores. Extraction of DNA from joining analysis of Jaccard’s coefficient of biotic organisms to reproduce successfully cultured mycelia derived from single spores band matching). Furthermore, ascospore in harsh environments. gave 218–263 randomly amplified poly- progeny from different ascomata on ‘single’ Most lichenized fungi produce abundant morphic DNA (RAPD) markers4 for each thalli of O. -

A Higher-Level Phylogenetic Classification of the Fungi
mycological research 111 (2007) 509–547 available at www.sciencedirect.com journal homepage: www.elsevier.com/locate/mycres A higher-level phylogenetic classification of the Fungi David S. HIBBETTa,*, Manfred BINDERa, Joseph F. BISCHOFFb, Meredith BLACKWELLc, Paul F. CANNONd, Ove E. ERIKSSONe, Sabine HUHNDORFf, Timothy JAMESg, Paul M. KIRKd, Robert LU¨ CKINGf, H. THORSTEN LUMBSCHf, Franc¸ois LUTZONIg, P. Brandon MATHENYa, David J. MCLAUGHLINh, Martha J. POWELLi, Scott REDHEAD j, Conrad L. SCHOCHk, Joseph W. SPATAFORAk, Joost A. STALPERSl, Rytas VILGALYSg, M. Catherine AIMEm, Andre´ APTROOTn, Robert BAUERo, Dominik BEGEROWp, Gerald L. BENNYq, Lisa A. CASTLEBURYm, Pedro W. CROUSl, Yu-Cheng DAIr, Walter GAMSl, David M. GEISERs, Gareth W. GRIFFITHt,Ce´cile GUEIDANg, David L. HAWKSWORTHu, Geir HESTMARKv, Kentaro HOSAKAw, Richard A. HUMBERx, Kevin D. HYDEy, Joseph E. IRONSIDEt, Urmas KO˜ LJALGz, Cletus P. KURTZMANaa, Karl-Henrik LARSSONab, Robert LICHTWARDTac, Joyce LONGCOREad, Jolanta MIA˛ DLIKOWSKAg, Andrew MILLERae, Jean-Marc MONCALVOaf, Sharon MOZLEY-STANDRIDGEag, Franz OBERWINKLERo, Erast PARMASTOah, Vale´rie REEBg, Jack D. ROGERSai, Claude ROUXaj, Leif RYVARDENak, Jose´ Paulo SAMPAIOal, Arthur SCHU¨ ßLERam, Junta SUGIYAMAan, R. Greg THORNao, Leif TIBELLap, Wendy A. UNTEREINERaq, Christopher WALKERar, Zheng WANGa, Alex WEIRas, Michael WEISSo, Merlin M. WHITEat, Katarina WINKAe, Yi-Jian YAOau, Ning ZHANGav aBiology Department, Clark University, Worcester, MA 01610, USA bNational Library of Medicine, National Center for Biotechnology Information, -

Diversity and Roles of Mycorrhizal Fungi in the Bee Orchid Ophrys Apifera
Diversity and Roles of Mycorrhizal Fungi in the Bee Orchid Ophrys apifera By Wazeera Rashid Abdullah April 2018 A Thesis submitted to the University of Liverpool in fulfilment of the requirement for the degree of Doctor in Philosophy Table of Contents Page No. Acknowledgements ............................................................................................................. xiv Abbreviations ............................................................................ Error! Bookmark not defined. Abstract ................................................................................................................................... 2 1 Chapter one: Literature review: ........................................................................................ 3 1.1 Mycorrhiza: .................................................................................................................... 3 1.1.1Arbuscular mycorrhiza (AM) or Vesicular-arbuscular mycorrhiza (VAM): ........... 5 1.1.2 Ectomycorrhiza: ...................................................................................................... 5 1.1.3 Ectendomycorrhiza: ................................................................................................ 6 1.1.4 Ericoid mycorrhiza, Arbutoid mycorrhiza, and Monotropoid mycorrhiza: ............ 6 1.1.5 Orchid mycorrhiza: ................................................................................................. 7 1.1.5.1 Orchid mycorrhizal interaction: ...................................................................... -

The Classification of Lower Organisms
The Classification of Lower Organisms Ernst Hkinrich Haickei, in 1874 From Rolschc (1906). By permission of Macrae Smith Company. C f3 The Classification of LOWER ORGANISMS By HERBERT FAULKNER COPELAND \ PACIFIC ^.,^,kfi^..^ BOOKS PALO ALTO, CALIFORNIA Copyright 1956 by Herbert F. Copeland Library of Congress Catalog Card Number 56-7944 Published by PACIFIC BOOKS Palo Alto, California Printed and bound in the United States of America CONTENTS Chapter Page I. Introduction 1 II. An Essay on Nomenclature 6 III. Kingdom Mychota 12 Phylum Archezoa 17 Class 1. Schizophyta 18 Order 1. Schizosporea 18 Order 2. Actinomycetalea 24 Order 3. Caulobacterialea 25 Class 2. Myxoschizomycetes 27 Order 1. Myxobactralea 27 Order 2. Spirochaetalea 28 Class 3. Archiplastidea 29 Order 1. Rhodobacteria 31 Order 2. Sphaerotilalea 33 Order 3. Coccogonea 33 Order 4. Gloiophycea 33 IV. Kingdom Protoctista 37 V. Phylum Rhodophyta 40 Class 1. Bangialea 41 Order Bangiacea 41 Class 2. Heterocarpea 44 Order 1. Cryptospermea 47 Order 2. Sphaerococcoidea 47 Order 3. Gelidialea 49 Order 4. Furccllariea 50 Order 5. Coeloblastea 51 Order 6. Floridea 51 VI. Phylum Phaeophyta 53 Class 1. Heterokonta 55 Order 1. Ochromonadalea 57 Order 2. Silicoflagellata 61 Order 3. Vaucheriacea 63 Order 4. Choanoflagellata 67 Order 5. Hyphochytrialea 69 Class 2. Bacillariacea 69 Order 1. Disciformia 73 Order 2. Diatomea 74 Class 3. Oomycetes 76 Order 1. Saprolegnina 77 Order 2. Peronosporina 80 Order 3. Lagenidialea 81 Class 4. Melanophycea 82 Order 1 . Phaeozoosporea 86 Order 2. Sphacelarialea 86 Order 3. Dictyotea 86 Order 4. Sporochnoidea 87 V ly Chapter Page Orders. Cutlerialea 88 Order 6. -

Towards an Integrated Phylogenetic Classification of the Tremellomycetes
http://www.diva-portal.org This is the published version of a paper published in Studies in mycology. Citation for the original published paper (version of record): Liu, X., Wang, Q., Göker, M., Groenewald, M., Kachalkin, A. et al. (2016) Towards an integrated phylogenetic classification of the Tremellomycetes. Studies in mycology, 81: 85 http://dx.doi.org/10.1016/j.simyco.2015.12.001 Access to the published version may require subscription. N.B. When citing this work, cite the original published paper. Permanent link to this version: http://urn.kb.se/resolve?urn=urn:nbn:se:nrm:diva-1703 available online at www.studiesinmycology.org STUDIES IN MYCOLOGY 81: 85–147. Towards an integrated phylogenetic classification of the Tremellomycetes X.-Z. Liu1,2, Q.-M. Wang1,2, M. Göker3, M. Groenewald2, A.V. Kachalkin4, H.T. Lumbsch5, A.M. Millanes6, M. Wedin7, A.M. Yurkov3, T. Boekhout1,2,8*, and F.-Y. Bai1,2* 1State Key Laboratory for Mycology, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, PR China; 2CBS Fungal Biodiversity Centre (CBS-KNAW), Uppsalalaan 8, Utrecht, The Netherlands; 3Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures, Braunschweig 38124, Germany; 4Faculty of Soil Science, Lomonosov Moscow State University, Moscow 119991, Russia; 5Science & Education, The Field Museum, 1400 S. Lake Shore Drive, Chicago, IL 60605, USA; 6Departamento de Biología y Geología, Física y Química Inorganica, Universidad Rey Juan Carlos, E-28933 Mostoles, Spain; 7Department of Botany, Swedish Museum of Natural History, P.O. Box 50007, SE-10405 Stockholm, Sweden; 8Shanghai Key Laboratory of Molecular Medical Mycology, Changzheng Hospital, Second Military Medical University, Shanghai, PR China *Correspondence: F.-Y. -

12 Tremellomycetes and Related Groups
12 Tremellomycetes and Related Groups 1 1 2 1 MICHAEL WEIß ,ROBERT BAUER ,JOSE´ PAULO SAMPAIO ,FRANZ OBERWINKLER CONTENTS I. Introduction I. Introduction ................................ 00 A. Historical Concepts. ................. 00 Tremellomycetes is a fungal group full of con- B. Modern View . ........................... 00 II. Morphology and Anatomy ................. 00 trasts. It includes jelly fungi with conspicuous A. Basidiocarps . ........................... 00 macroscopic basidiomes, such as some species B. Micromorphology . ................. 00 of Tremella, as well as macroscopically invisible C. Ultrastructure. ........................... 00 inhabitants of other fungal fruiting bodies and III. Life Cycles................................... 00 a plethora of species known so far only as A. Dimorphism . ........................... 00 B. Deviance from Dimorphism . ....... 00 asexual yeasts. Tremellomycetes may be benefi- IV. Ecology ...................................... 00 cial to humans, as exemplified by the produc- A. Mycoparasitism. ................. 00 tion of edible Tremella fruiting bodies whose B. Tremellomycetous Yeasts . ....... 00 production increased in China alone from 100 C. Animal and Human Pathogens . ....... 00 MT in 1998 to more than 250,000 MT in 2007 V. Biotechnological Applications ............. 00 VI. Phylogenetic Relationships ................ 00 (Chang and Wasser 2012), or extremely harm- VII. Taxonomy................................... 00 ful, such as the systemic human pathogen Cryp- A. Taxonomy in Flow -

Sex, Outcrossing and Mating Types: Unsolved Questions in Fungi and Beyond
doi: 10.1111/j.1420-9101.2012.02495.x REVIEW Sex, outcrossing and mating types: unsolved questions in fungi and beyond S. BILLIARD*, M. LO´ PEZ-VILLAVICENCIO ,M.E.HOODà &T.GIRAUD§– *Laboratoire de Ge´ne´tique et Evolution des Populations Ve´ge´tales, UMR CNRS 8016, Universite´ des Sciences et Technologies de Lille – Lille1, Villeneuve d’Ascq Cedex, France Origine, Structure, Evolution de la Biodiversite´, UMR 7205 CNRS-MNHN, Muse´um National d’Histoire Naturelle, Paris Cedex, France àDepartment of Biology, McGuire Life Sciences Building, Amherst College, Amherst, MA, USA §Ecologie, Syste´matique et Evolution, Universite´ Paris-Sud, Orsay cedex, France –Ecologie, Syste´matique et Evolution, CNRS, Orsay cedex, France Keywords: Abstract ascomycete; Variability in the way organisms reproduce raises numerous, and still asexual reproduction; unsolved, questions in evolutionary biology. In this study, we emphasize that basidiomycete; fungi deserve a much greater emphasis in efforts to address these questions breeding systems; because of their multiple advantages as model eukaryotes. A tremendous diploid selfing; diversity of reproductive modes and mating systems can be found in fungi, gametophytic selfing; with many evolutionary transitions among closely related species. In addition, haploid selfing; fungi show some peculiarities in their mating systems that have received little mating systems; attention so far, despite the potential for providing insights into important oomycetes; evolutionary questions. In particular, selfing can occur at the haploid stage in sexual reproduction. addition to the diploid stage in many fungi, which is generally not possible in animals and plants but has a dramatic influence upon the structure of genetic systems. Fungi also present several advantages that make them tractable models for studies in experimental evolution.