Two Genetic Variants Associated with Plantar Fascial Disorders
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Acral Manifestations of Soft Tissue Tumors Kristen M
Clinics in Dermatology (2017) 35,85–98 Acral manifestations of soft tissue tumors Kristen M. Paral, MD, Vesna Petronic-Rosic, MD, MSc⁎ Section of Dermatology, University of Chicago Pritzker School of Medicine, Chicago, IL Abstract This group of biologically diverse entities is united by topographic localization to the hands and feet. Categorizing tumors by body site narrows the differential into a short list of possibilities that can facil- itate accurate and rapid diagnosis. The goal of this review is to provide a practical approach to soft tissue tumors of acral locations for clinicians, pathologists, and researchers alike. What ensues in the following text is that tight coupling of the clinical picture and histopathologic findings should produce the correct diagno- sis, or at least an abbreviated differential. The salient clinicopathologic, immunohistochemical, and molec- ular features are presented alongside current treatment recommendations for each entity. © 2017 Elsevier Inc. All rights reserved. Introduction actin (SMA) and are deemed “myofibroblasts.”1 The tumors under this heading express combinations of CD34, FXIIIa, fi The entities presented herein are categorized on the basis of and SMA. The synthesis of collagen by broblasts translates fi fi morphogenesis (where possible) and by biologic potential as to a brous consistency that clinically imparts a rm texture benign, intermediate, and malignant neoplasms. on palpation, and, macroscopically, a gray-white or white-tan cut surface. The entities discussed next have no metastatic po- tential; that is, simple excision is adequate. Fibrous and related tissues: Benign lesions Fibroma of tendon sheath fi fl The ontogenetic classi cation of benign lesions re ects appar- Also known as tenosynovial fibroma, compared with other fi fi fi ent broblastic or broblast-like morphogenesis. -

The Best Diagnosis Is
Dermatopathology Diagnosis Superficial Plantar Fibromatosis The best diagnosis is: H&E, original magnification 40. a. dermatofibroma b. keloid CUTISc. neurofibroma d. nodular fasciitis Do Note. superficialCopy plantar fibromatosis H&E, original magnification 400. PLEASE TURN TO PAGE 225 FOR DERMATOPATHOLOGY DIAGNOSIS DISCUSSION Luke Lennox, BA; Anna Li, BS; Thomas N. Helm, MD Mr. Lennox and Dr. Helm are from the Department of Dermatology, University at Buffalo, The State University of New York. Ms. Li is from the Department of Dermatology, Ross University, Dominica, West Indies. The authors report no conflict of interest. Correspondence: Thomas N. Helm, MD, Dermatopathology Laboratory, 6255 Sheridan Dr, Building B, Ste 208, Williamsville, NY 14221 ([email protected]). 220 CUTIS® WWW.CUTIS.COM Copyright Cutis 2013. No part of this publication may be reproduced, stored, or transmitted without the prior written permission of the Publisher. Dermatopathology Diagnosis Discussion Superficial Plantar Fibromatosis lantar fibromatosis typically presents as firm represents a reactive proliferation of spindle cells most plaques or nodules on the plantar surface of the often encountered on the extremities of young adults. Pfoot.1 The process is caused by a proliferation of Spindle cells are loosely arranged in a mucinous fibroblasts and collagen and has been associated with stroma and are not circumscribed (tissue culture ap- trauma, liver disease, diabetes mellitus, epilepsy, and pearance). Vesicular nuclei are encountered, but there alcoholism.2 Unlike the fibromatoses associated with is no remarkable nuclear pleomorphism. Extravasated Gardner syndrome, superficial plantar fibromatosis has not been associated with abnormalities in the ade- nomatous polyposis coli gene or with the -catenin gene.3,4 Lesions typically present in middle-aged or elderly individuals and involve the medial plantar fas- cia. -

A Follow-Up of Patient Reported Outcomes in Chronic Plantar Heel Pain Participants Treated with Graston Technique: a Mixed Methods Approach
University of Northern Iowa UNI ScholarWorks Dissertations and Theses @ UNI Student Work 2016 A follow-up of patient reported outcomes in chronic plantar heel pain participants treated with Graston Technique: A mixed methods approach Troy Richard Garrett University of Northern Iowa Let us know how access to this document benefits ouy Copyright ©2016 Troy Richard Garrett Follow this and additional works at: https://scholarworks.uni.edu/etd Part of the Physical Therapy Commons, and the Physiotherapy Commons Recommended Citation Garrett, Troy Richard, "A follow-up of patient reported outcomes in chronic plantar heel pain participants treated with Graston Technique: A mixed methods approach" (2016). Dissertations and Theses @ UNI. 344. https://scholarworks.uni.edu/etd/344 This Open Access Dissertation is brought to you for free and open access by the Student Work at UNI ScholarWorks. It has been accepted for inclusion in Dissertations and Theses @ UNI by an authorized administrator of UNI ScholarWorks. For more information, please contact [email protected]. Copyright by TROY RICHARD GARRETT 2016 All Rights Reserved A FOLLOW-UP OF PATIENT REPORTED OUTCOMES IN CHRONIC PLANTAR HEEL PAIN PARTICIPANTS TREATED WITH GRASTON TECHNIQUE: A MIXED METHODS APPROACH An Abstract of a Dissertation Submitted in Partial Fulfillment of the Requirements for the Degree Doctor of Education Approved: ____________________________________ Dr. Peter J. Neibert, Committee Chair ____________________________________ Dr. Kavita R. Dhanwada Dean of the Graduate College Troy Richard Garrett University of Northern Iowa December 2016 ABSTRACT Chronic Plantar Heel Pain (CPHP), commonly known as “plantar fasciitis,” is a condition that is estimated to affect 10% of the American population. -

A Pilot Study of a Novel Treatment Method for Refractory Painful Plantar Fibromas
Open Access Austin Journal of Orthopedics & Rheumatology Research Article A Pilot Study of a Novel Treatment Method for Refractory Painful Plantar Fibromas Mihir M. Patel*, Sahvan M. Patel, Sia S. Patel and Jacob Daynes Abstract Department of Orthopedic Surgery, OrthoIndy, USA Introduction: Painful plantar fibromas may make ambulation difficult *Corresponding author: Mihir M Patel, Department for patients. They may recur and can make shoe wear difficult to purchase. of Orthopedic Surgery, OrthoIndy 8450 Northwest Blvd, Treatment modalities may include conservative care, modified shoe wear, Indianapolis, Indiana 46278, USA injections and orthotics. Surgical treatment may include open excision. The TX1A probe by Tenex Inc may be a useful alternative treatment modality for Received: March 18, 2015; Accepted: April 22, 2015; these painful lesions. Published: April 22, 2015 Materials and Methods: From 2011 through 2014, eight patients who had painful plantar fibromas elected to undergo definitive treatment for the fibromas. They all had had conservative care including modifications of shoes and either over the counter orthotics or custom orthotics. They all had advanced imaging tests (MRI or CT) preoperatively to help delineate the lesions. The definitive treatment utilized arthroscopy and the TX1A probe and was performed on an outpatient basis. Results: Preoperative AOFAS score was 30.8 (20-36). Postoperative AOFAS score was 90.1 (85-92). Average time of resolution of symptoms was 63.5 days (30-112) and average follow-up was 2.5 years. One patient had another lesion become painful but it was in a different location than the index operation. No others have had a recurrence to date. -

Plantar Fasciitis Standard of Care
BRIGHAM AND WOMEN’S HOSPITAL Department of Rehabilitation Services Physical Therapy Standard of Care: Plantar Fasciitis Case Type / Diagnosis: (diagnosis specific, impairment/ dysfunction specific) ICD9 - 728.71 plantar fibromatosis Plantar fasciitis is an inflammatory condition that occurs as a result of overstressing the plantar fascia. It is the most common cause of inferior heel pain and has been diagnosed in patients from the ages of 8-80. 1-4 Plantar fasciitis affects approximately 10% of the population and is more commonly found in middle-aged women and younger male runners.2, 3, 5 A majority of the patients diagnosed with this inflammatory condition are over the age of 40.3 Bilateral symptoms can occur in 20-30% of those diagnosed with plantar fasciitis.6 However in these cases it is important to rule out other systemic processes such as rheumatoid arthritis, systemic lupus erythematosus, Reiter’s disease, gout and ankylosing spondylitis.3, 6 The primary symptom of plantar fasciitis is pain in the heel when the patient first rises in the morning and when the plantar fascia is palpated over it’s origin at the medial calcaneal tuberosity.2-4, 6, 7 The plantar fascia (aponeurosis) is a thick fibrous band of connective tissue that originates at the medial and lateral tuberosities of the calcaneus. It runs longitudinally and has 3 portions, medial, central, and lateral. At the midfoot level, the fascia divides into 5 separate bands, which blend with the flexor tendon sheaths and transverse metatarsal ligaments and attach at the base of each proximal phalange. The central portion of the plantar fascia is the largest section. -
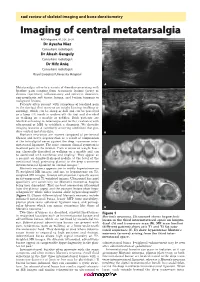
Imaging of Central Metatarsalgia
rad review of skeletal imaging and bone densitometry Imaging of central metatarsalgia RAD Magazine, 45, 528, 29-30 Dr Ayesha Niaz Consultant radiologist Dr Akash Ganguly Consultant radiologist Dr Hifz Aniq Consultant radiologist Royal Liverpool University Hospital A Metatarsalgia refers to a variety of disorders presenting with forefoot pain ranging from traumatic lesions (acute or chronic repetitive), inflammatory and infective disorders, non-neoplastic soft tissue lesions, and benign tumours to malignant lesions. Patients often present with symptoms of localised pain in the forefoot that worsens on weight bearing (walking or running), which can be sharp or dull and can be perceived as a lump felt inside or underneath the foot and described as walking on a marble or pebbles. Such patients are labelled as having metatarsalgia and further evaluated with ultrasound or MRI to establish a diagnosis. We describe imaging features of commonly occurring conditions that pro- duce central metatarsalgia. B Morton’s neuromas are masses composed of perineural fibrosis and nerve degeneration as a result of compression of the interdigital nerve against the deep transverse inter- metatarsal ligament. The most common clinical symptom is localised pain in the forefoot. Pain is worse on weight bear- ing, classically described as walking on a marble and can be associated with numbness and tingling.1 They appear as a peanut- or dumbbell-shaped nodule at the level of the metatarsal head, projecting plantar to the deep transverse intermetatarsal ligament on coronal images.2 Morton’s neuroma appears iso- to mildly hyperintense on C T1-weighted MR images and iso- to hypointense on T2- weighted MR images. -

Synovial Sarcoma- an Unusual Cause of Heel Pain
Malaysian Orthopaedic Journal 2008 Vol 2 No 2 MY Norhamdan, et al Synovial Sarcoma- An unusual cause of Heel Pain MY Norhamdan, MS (Ortho), Y Shahril, MS (Ortho), O Masbah, MS (Ortho), Siti Aishah MA*, MIAC Department of Orthopaedics & Traumatology, UKM Medical Centre, Kuala Lumpur, Malaysia * Department of Pathology, UKM Medical Centre, Kuala Lumpur, Malaysia ABSTRACT imaging revealed a well-circumscribed heterogeneous lesion, isointense to muscle on T(1)-weighted image and We report a case of 29-year-old female who presented with hyperintense on T(2) image indicating a benign lesion right heel pain that worsened over a period of two years. The (Figure 2). Due to close proximity to the posterior tibial onset of pain was followed by swelling at the medial aspect nerve, the patient was diagnosed with neuroma. of right ankle. She was initially treated for plantar fasciitis with multiple steroid injections over the heel. Subsequent The patient underwent excision biopsy and intra-operatively MRI revealed a well-defined heterogeneous lesion in the lesion was noted to be a soft, friable mass encasing the continuity with the medial plantar nerve. Excision biopsy medial plantar nerve (Figure 3). Histopathological was performed and histopathological evaluation revealed evaluation revealed monophasic synovial sarcoma as monophasic synovial sarcoma. The patient subsequently evidenced by spindle-shaped and neoplastic cells. underwent wide resection and free tissue transfer followed by Imunohistochemically the neoplastic cells were positive for radiotherapy and chemotherapy. This case highlights an vimentin and focally positive for EMA, CK, and CD 99 but unusual site and presentation of synovial sarcoma which led negative for CD 34, CD 31, desmin, myoglobin, smooth to delayed diagnosis and treatment. -

EXTRACORPOREAL SHOCK WAVE THERAPY (ESWT) Policy Number: SUR036 Effective Date: April 1, 2019
UnitedHealthcare® Commercial Medical Policy EXTRACORPOREAL SHOCK WAVE THERAPY (ESWT) Policy Number: SUR036 Effective Date: April 1, 2019 Table of Contents Page COVERAGE RATIONALE ............................................. 1 CENTERS FOR MEDICARE AND MEDICAID SERVICES APPLICABLE CODES ................................................. 1 (CMS) .................................................................. 11 DESCRIPTION OF SERVICES ...................................... 2 REFERENCES ........................................................ 11 CLINICAL EVIDENCE ................................................ 2 POLICY HISTORY/REVISION INFORMATION ............... 14 U.S. FOOD AND DRUG ADMINISTRATION (FDA) ......... 10 INSTRUCTIONS FOR USE ........................................ 14 COVERAGE RATIONALE Extracorporeal shock wave therapy (ESWT), whether low energy, high energy or radial wave, is unproven and not medically necessary for all indications, including but not limited to the treatment of: Achilles tendonitis Calcaneal spur Calcific tendonitis of the shoulder (rotator cuff) Chronic plantar fasciitis (including plantar fibromatosis and plantar nerve lesion) Delayed or nonunion of fractures Hammer toe Lateral epicondylitis (tennis elbow) Medial epicondylitis (golfers elbow) Tenosynovitis of the foot or ankle Tibialis tendinitis Wounds including ulcers The available evidence regarding the efficacy of ESWT is conflicting. There is insufficient evidence regarding the durability of the treatment effects of ESWT. Patient selection -

Case: 5:12-Cv-01888-CAB Doc #: 13 Filed: 04/29/13 1 of 30. Pageid
Case: 5:12-cv-01888-CAB Doc #: 13 Filed: 04/29/13 1 of 30. PageID #: <pageID> IN THE UNITED STATES DISTRICT COURT FOR THE NORTHERN DISTRICT OF OHIO EASTERN DIVISION SUSAN MARIE LUCKY, ) ) CASE NO. 5:12CV1888 Plaintiff, ) ) v. ) JUDGE CHRISTOPHER A. BOYKO ) ) MAGISTRATE JUDGE GREG WHITE ) MICHAEL J. ASTRUE, ) Commissioner of Social Security ) REPORT & RECOMMENDATION ) Defendant. ) Plaintiff Susan Marie Lucky (“Lucky”) challenges the final decision of the Commissioner of Social Security, Michael J. Astrue (“Commissioner”), denying her claim for a period of disability (“POD”) and disability insurance benefits (“DIB”) under Title II of the Social Security Act (“Act”), 42 U.S.C. §§ 416(i) and 423 et seq. This matter is before the Court pursuant to 42 U.S.C. § 405(g) and Local Rule 72.2(b). For the reasons set forth below, it is recommended that the final decision of the Commissioner be affirmed in part, vacated in part, and the case remanded. I. Procedural History On October 22, 2007, Lucky filed an application for POD and DIB, alleging a disability Case: 5:12-cv-01888-CAB Doc #: 13 Filed: 04/29/13 2 of 30. PageID #: <pageID> onset date of October 1, 2005 and claiming she was disabled due to a variety of conditions, including fibromyalgia, fibromatosis, colitis, inflammatory arthritis, tendonitis, and carpal tunnel syndrome. Her application was denied both initially and upon reconsideration. On May 13, 2010, Lucky submitted a letter to the Administration requesting a closed period of benefits between October 1, 2005 to December 1, 2007, and ongoing disability since November 1, 2009. -

Myxochondroid Metaplasia of the Plantar Foot
Modern Pathology (2013) 26, 1561–1567 & 2013 USCAP, Inc. All rights reserved 0893-3952/13 $32.00 1561 Myxochondroid metaplasia of the plantar foot: a distinctive pseudoneoplastic lesion resembling nuchal fibrocartilaginous pseudotumor and the equine digital cushion Wonwoo Shon and Andrew L Folpe Department of Laboratory Medicine and Pathology, Mayo Clinic, Rochester, MN, USA Cartilaginous tumors of soft tissue are uncommon, with benign chondromas of soft parts greatly outnumbering rare soft-tissue chondrosarcomas. Over the past several years, we have seen in consultation a distinctive, benign-appearing chondroid soft-tissue lesion of the plantar foot that differs in a number of respects from chondroma of soft parts. Herein we report our experience with this distinctive lesion. A retrospective review of all cases from the foot in our soft-tissue consultation and institutional surgical pathology archives identified 9 similar cases, most often previously coded as ‘fibroconnective tissue with chondroid metaplasia’. Six cases were submitted in consultation due to concern for a neoplastic process, in particular chondroma of soft parts or fibro-osseous pseudotumor of the digits. The patients were 4 young males (age range 8–16 years, mean 11.5 years) and 5 older patients, including 4 women and 1 man (age range 34–78 years, mean 56.4 years). All cases occurred in the subcutaneous plantar soft tissues of the feet, including four cases confined to the toes, and presented as non-specific, variably painful masses. Radiographic studies, available in six cases, did not show any evidence of bone involvement. Histologically, the lesions were characterized by a partially circumscribed, variably cellular proliferation of bland fibroblastic cells in a fibromyxoid background in areas showing distinct stromal basophilia and a chondroid appearance. -

2014 Slide Library Case Summary Questions & Answers With
2014 Slide Library Case Summary Questions & Answers with Discussions 51st Annual Meeting November 6-9, 2014 Chicago Hilton & Towers Chicago, Illinois The American Society of Dermatopathology ARTHUR K. BALIN, MD, PhD, FASDP FCAP, FASCP, FACP, FAAD, FACMMSCO, FASDS, FAACS, FASLMS, FRSM, AGSF, FGSA, FACN, FAAA, FNACB, FFRBM, FMMS, FPCP ASDP REFERENCE SLIDE LIBRARY November 2014 Dear Fellows of the American Society of Dermatopathology, The American Society of Dermatopathology would like to invite you to submit slides to the Reference Slide Library. At this time there are over 9300 slides in the library. The collection grew 2% over the past year. This collection continues to grow from member’s generous contributions over the years. The slides are appreciated and are here for you to view at the Sally Balin Medical Center. Below are the directions for submission. Submission requirements for the American Society of Dermatopathology Reference Slide Library: 1. One H & E slide for each case (if available) 2. Site of biopsy 3. Pathologic diagnosis Not required, but additional information to include: 1. Microscopic description of the slide illustrating the salient diagnostic points 2. Clinical history and pertinent laboratory data, if known 3. Specific stain, if helpful 4. Clinical photograph 5. Additional note, reference or comment of teaching value Teaching sets or collections of conditions are especially useful. In addition, infrequently seen conditions are continually desired. Even a single case is helpful. Usually, the written submission requirement can be fulfilled by enclosing a copy of the pathology report prepared for diagnosis of the submitted case. As a guideline, please contribute conditions seen with a frequency of less than 1 in 100 specimens. -

First-Time Report
www.medigraphic.org.mx Medicina Cutánea Ibero-Latino-Americana Localizador: 17057 Ledderhose’s disease associated with Dupuytren’s disease in a patient with acquired immunodeficiency syndrome: first-time report Doença de Ledderhose associada à doença de Dupuytren em paciente com síndrome da imunodefi ciência humana: primeiro relato Raquel Nardelli de Araujo,* Renata Marcarini,* Aaron Alarcon Novillo,‡ Daniela Alves Pereira Antelo§ Key words: Plantar fibromatosis, ABSTRACT RESUMO Ledderhose’s disease, acquired The plantar fibromatosis (PF), also known as Ledderhose’s A fibromatose plantar (FP), também conhecida como doença de immunodeficiency disease, is characterized by affecting the plantar aponeurosis. Ledderhose, é uma doença caracterizada por acometer a syndrome. In a general way, it is a benign fibrous condition still quite aponeurose plantar. Trata-se de uma condição fibrosa benigna unclear in its various aspects, such as its etiology and treatment. ainda pouco clara nos seus diversos aspectos, como a etiologia Palavras-chave: To date, it has not been described in association with AIDS. e o tratamento. Descrevemos o caso de um paciente de 45 anos Fibromatose Our patient presented PF previously hospitalization due to do sexo masculino portador de síndrome da imunodeficiência plantar, doença de occurrence of many opportunistic infections due to AIDS at an adquirida em fase avançada, associado à doença de Dupuytren Ledderhose, síndrome advanced stage. We have described the case of a 45 year-old (DD) que evoluiu com o surgimento da FP bilateral. O tratamento da imunodeficiência male patient with acquired immunodeficiency syndrome at a foi conservador, pois as lesões plantares eram assintomáticas adquirida. late stage, associated with Dupuytren’s disease (DD) and the e o risco de recorrência com medidas invasivas é elevado.