At Least Seven Distinct Rotavirus Genotype Constellations in Bats with Evidence Of
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
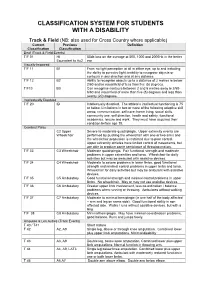
Disability Classification System
CLASSIFICATION SYSTEM FOR STUDENTS WITH A DISABILITY Track & Field (NB: also used for Cross Country where applicable) Current Previous Definition Classification Classification Deaf (Track & Field Events) T/F 01 HI 55db loss on the average at 500, 1000 and 2000Hz in the better Equivalent to Au2 ear Visually Impaired T/F 11 B1 From no light perception at all in either eye, up to and including the ability to perceive light; inability to recognise objects or contours in any direction and at any distance. T/F 12 B2 Ability to recognise objects up to a distance of 2 metres ie below 2/60 and/or visual field of less than five (5) degrees. T/F13 B3 Can recognise contours between 2 and 6 metres away ie 2/60- 6/60 and visual field of more than five (5) degrees and less than twenty (20) degrees. Intellectually Disabled T/F 20 ID Intellectually disabled. The athlete’s intellectual functioning is 75 or below. Limitations in two or more of the following adaptive skill areas; communication, self-care; home living, social skills, community use, self direction, health and safety, functional academics, leisure and work. They must have acquired their condition before age 18. Cerebral Palsy C2 Upper Severe to moderate quadriplegia. Upper extremity events are Wheelchair performed by pushing the wheelchair with one or two arms and the wheelchair propulsion is restricted due to poor control. Upper extremity athletes have limited control of movements, but are able to produce some semblance of throwing motion. T/F 33 C3 Wheelchair Moderate quadriplegia. Fair functional strength and moderate problems in upper extremities and torso. -

Ifds Functional Classification System & Procedures
IFDS FUNCTIONAL CLASSIFICATION SYSTEM & PROCEDURES MANUAL 2009 - 2012 Effective – 1 January 2009 Originally Published – March 2009 IFDS, C/o ISAF UK Ltd, Ariadne House, Town Quay, Southampton, Hampshire, SO14 2AQ, GREAT BRITAIN Tel. +44 2380 635111 Fax. +44 2380 635789 Email: [email protected] Web: www.sailing.org/disabled 1 Contents Page Introduction 5 Part A – Functional Classification System Rules for Sailors A1 General Overview and Sailor Evaluation 6 A1.1 Purpose 6 A1.2 Sailing Functions 6 A1.3 Ranking of Functional Limitations 6 A1.4 Eligibility for Competition 6 A1.5 Minimum Disability 7 A2 IFDS Class and Status 8 A2.1 Class 8 A2.2 Class Status 8 A2.3 Master List 10 A3 Classification Procedure 10 A3.0 Classification Administration Fee 10 A3.1 Personal Assistive Devices 10 A3.2 Medical Documentation 11 A3.3 Sailors’ Responsibility for Classification Evaluation 11 A3.4 Sailor Presentation for Classification Evaluation 12 A3.5 Method of Assessment 12 A3.6 Deciding the Class 14 A4 Failure to attend/Non Co-operation/Misrepresentation 16 A4.1 Sailor Failure to Attend Evaluation 16 A4.2 Non Co-operation during Evaluation 16 A4.3 International Misrepresentation of Skills and/or Abilities 17 A4.4 Consequences for Sailor Support Personnel 18 A4.5 Consequences for Teams 18 A5 Specific Rules for Boat Classes 18 A5.1 Paralympic Boat Classes 18 A5.2 Non-Paralympic Boat Classes 19 Part B – Protest and Appeals B1 Protest 20 B1.1 General Principles 20 B1.2 Class Status and Protest Opportunities 21 B1.3 Parties who may submit a Classification Protest -

Cam Switches
Cam switches 10 Cam operated switches 11 General characteristics 12 Reference system 13 Technical data 14 Dimensions 18 Auxiliary contacts 19 Mounting possibilities 20 Mounting schemes 22 Special mountings 25 Accessories 31 Enclosures 32 Standard electrical schemes 46 Special diagrams 48 Main switches with undervoltage release 50 Discrepancy switches Cam switches Cam operated switches A5 cam operated switches have General characteristics been designed to operate as switch-disconnectors, main • Permits any electrical diagrams oxidation of their ferrous switches, load break switches, and makes the number of components. changeover switches, motor possible diagrams limitless. • High number of mechanical control switches,… • Abrupt breaking mechanism operations. with 30, 45, 60 o 90° positions, • According to RoHS standard. According to Standards according to diagram and requirements. • IEC 60947-3 • Contact decks made of self- • UL 508 extinguishing polyester reinforced with glass fi ber. • Silver alloy low resistance contacts with high arcing and welding characteristics. • Connection with protected cable clamps until 125A. • Electrolytic treatment against 10 low voltage electrical material Serie A5 low voltage electrical material General characteristics Simple "click" front plate fi xing Front plate designed for Internal and easy fi xing by simple push-in on the external links mounting plate Factory assembled links. Insulated external links protect against direct contact on live parts Metalic shaft Marking Optimal mechanical eff ort transmission -

Surgical and Non-Surgical Management Strategies for Lateral Epicondylitis
J Orthop Spine Trauma. 2021 March; 7(1): 1-7. DOI: http://dx.doi.org/10.18502/jost.v7i1.5958 Review Article Surgical and Non-Surgical Management Strategies for Lateral Epicondylitis Meisam Jafari Kafiabadi 1, Amir Sabaghzadeh1, Farsad Biglari2, Amin Karami3, Mehrdad Sadighi 1,*, Adel Ebrahimpour4 1 Assistant Professor, Department of Orthopedic Surgery, Shohada Tajrish Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran 2 Orthopedic Surgeon, Department of Orthopedic Surgery, Shohada Tajrish Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran 3 Resident, Department of Orthopedic Surgery, Shohada Tajrish Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran 4 Professor, Department of Orthopedic Surgery, Shohada Tajrish Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran *Corresponding author: Mehrdad Sadighi; Department of Orthopedic Surgery, Shohada Tajrish Hospital, Shahid Beheshti University of Medical Sciences, Tehran, Iran. Tel: +98-9125754616, Email: [email protected] Received: 14 September 2020; Revised: 01 December 2020; Accepted: 12 January 2021 Abstract Lateral epicondylitis (LE) is one of the major causes of elbow pain. Despite being a self-limiting condition, its high incidence can cause a significant socioeconomic burden. Many treatment modalities have been proposed for the treatment, but the optimal strategy is still unknown. In this article, we discuss surgical and non-surgical strategies for the treatment of LE and address the research gaps. Keywords: Exercise Therapy; Lateral Epicondylitis; Tennis Elbow; Treatment Citation: Jafari Kafiabadi M, Sabaghzadeh A, Biglari F, Karami A, Sadighi M, Ebrahimpour A. Surgical and Non-Surgical Management Strategies for Lateral Epicondylitis. J Orthop Spine Trauma 2021; 7(1): 1-7. Background I-A. -

Nebraska Highway 12 Niobrara East & West
WWelcomeelcome Nebraska Highway 12 Niobrara East & West Draft Environmental Impact Statement and Section 404 Permit Application Public Open House and Public Hearing PPurposeurpose & NNeedeed What is the purpose of the N-12 project? - Provide a reliable roadway - Safely accommodate current and future traffi c levels - Maintain regional traffi c connectivity Why is the N-12 project needed? - Driven by fl ooding - Unreliable roadway, safety concerns, and interruption in regional traffi c connectivity Photo at top: N-12 with no paved shoulder and narrow lane widths Photo at bottom: Maintenance occurring on N-12 PProjectroject RRolesoles Tribal*/Public NDOR (Applicant) Agencies & County Corps » Provide additional or » Respond to the Corps’ » Provide technical input » Comply with Clean new information requests for information and consultation Water Act » Provide new reasonable » Develop and submit » Make the Section 7(a) » Comply with National alternatives a Section 404 permit decision (National Park Service) Environmental Policy Act application » Question accuracy and » Approvals and reviews » Conduct tribal adequacy of information » Planning, design, and that adhere to federal, consultation construction of Applied- state, or local laws and » Coordinate with NDOR, for Project requirements agencies, and public » Implementation of » Make a decision mitigation on NDOR’s permit application * Conducted through government-to-government consultation. AApplied-forpplied-for PProjectroject (Alternative A7 - Base of Bluff s Elevated Alignment) Attribute -
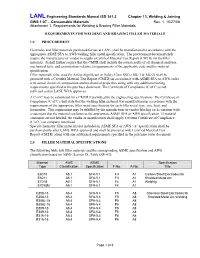
Requirements for Welding and Brazing Filler Materials
LANL Engineering Standards Manual ISD 341-2 Chapter 13, Welding & Joining GWS 1-07 – Consumable Materials Rev. 1, 10/27/06 Attachment 1, Requirements for Welding & Brazing Filler Materials REQUIREMENTS FOR WELDING AND BRAZING FILLER MATERIALS 1.0 PROCUREMENT Electrodes and filler materials purchased for use at LANL shall be manufactured in accordance with the appropriate ASME SFA or AWS welding filler metal specification. The procurement document shall require the manufacturer or vendor to supply a Certified Material Test Report (CMTR) for the filler materials. It shall further require that the CMTR shall include the actual results of all chemical analyses, mechanical tests, and examinations relative to requirements of the applicable code and the material specification. Filler materials to be used for Safety Significant or Safety Class SSCs (ML-1 & ML-2) shall be procured with a Certified Material Test Report (CMTR) in accordance with ASME SFA or AWS codes with actual chemical composition and mechanical properties along with any additional testing requirements specified in the purchase document. The Certificate of Compliance (C of C) is not sufficient unless LANL WPA approves. A C of C may be substituted for a CMTR if permitted by the engineering specification. The Certificate of Compliance (C of C) shall state that the welding filler material was manufactured in accordance with the requirements of the appropriate filler metal specification for each filler metal type, size, heat, and lot number. This requirement may be fulfilled by the manufacturer or vendor labeling each container with a statement that the material conforms to the appropriate ASME SFA or AWS specification. -

Temporary Assignment in Higher Classification
Commonwealth of Pennsylvania Governor's Office Subject: Number: Temporary Assignment in Higher 525.4 Amended Classification Date: By Direction of: May 3, 2013 Kelly Powell Logan, Secretary of Administration Contact Agency: Office of Administration, Office for Human Resources Management, Bureau of Classification and Compensation, Telephone 717.787.8838 This directive establishes policy, responsibilities, and procedures for controlling the use of temporary assignments in higher classifications and for processing payments to compensate employees who receive such assignments. Marginal dots are excluded due to major changes. 1. PURPOSE. To establish policy, responsibilities, and procedures for controlling the use of temporary assignments in higher classifications. 2. SCOPE. This directive applies to all employees in departments, boards, commissions, and councils (hereinafter referred to as “agencies”) under the Governor's jurisdiction temporarily assigned to the work of a position in a higher classification. 3. OBJECTIVES. a. Provide policy for the assignment of employees to temporarily work in a higher classification. b. Set forth eligibility criteria for receiving out-of-classification pay. c. Promulgate out-of-classification pay rules. d. Authorize the pay for employees temporarily working in a higher classification. Management Directive 525.4 Amended Page 1 of 8 4. DEFINITIONS. a. Employees' Regular Biweekly Salary/Hourly Rate. The biweekly salary/hourly rate the employee earns in his/her regular classification/position. b. Full day. Seven and one-half or eight hour day. Note that this may be any continuous seven and one-half or eight hour period including both hours within or outside of an employee’s regularly scheduled workday, or any combination thereof. -

US SAILING FUNCTIONAL CLASSIFICATION SYSTEM & PROCEDURES MANUAL 2013—2016 Effective—1 January 2013
[Type text ] [Type text ] [Type text ] US SAILING FUNCTIONAL CLASSIFICATION SYSTEM & PROCEDURES MANUAL 2013—2016 Effective—1 January 2013 Originally Published—May 2013 US Sailing Member of the PO Box 1260 15 Maritime Drive Portsmouth, RI 02871-0907 Phone: 1-401-683-0800 / Toll free: 1-800-USSAIL - (1-800-877-2451) Fax: 401-683-0840 E-Mail: [email protected] International Paralympic Committee Roger H S trube, MD – A pril 20 13 1 ROGE R H STRUBE, MD – APRIL 2013 [Type text ] [Type text ] [Type text ] Contents Introduction .................................................................................................................................................... 6 Appendix to Introduction – Glossary of Medical Terminology .......................................................................... 7 Joint Movement Definitions: ........................................................................................................................... 7 Neck ........................................................................................................................................................ 7 Shoulder .................................................................................................................................................. 7 Elbow ...................................................................................................................................................... 7 Wrist ...................................................................................................................................................... -
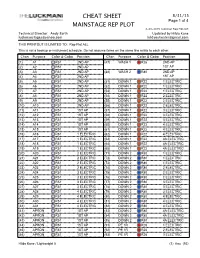
8-28-2015 Luckman Rep Plot.Lw5 Technical Director: Andy Barth Updated by Hilda Kane [email protected] [email protected]
CHEAT SHEET 8/31/15 Page 1 of 4 MAINSTAGE REP PLOT 8-28-2015 Luckman Rep Plot.lw5 Technical Director: Andy Barth Updated by Hilda Kane [email protected] [email protected] THIS PRINTOUT IS LIMITED TO: Rep Plot ALL This is not a hookup or instrument schedule. Do not assume items on the same line relate to each other. Chan Purpose Color & Gobo Position Chan Purpose Color & Gobo Position (1) A1 R51 2ND AP (47) WASH 1 R26 2ND AP (2) A2 R51 2ND AP 1ST AP (3) A3 R51 2ND AP (48) WASH 2 R80 2ND AP (4) A4 R51 2ND AP 1ST AP (5) A5 R51 2ND AP (51) DOWN 1 R22 1 ELECTRIC (6) A6 R51 2ND AP (52) DOWN 1 R22 1 ELECTRIC (7) A7 R51 2ND AP (53) DOWN 1 R22 1 ELECTRIC (8) A8 R51 2ND AP (54) DOWN 1 R22 2 ELECTRIC (9) A9 R51 2ND AP (55) DOWN 1 R22 2 ELECTRIC (10) A10 R51 2ND AP (56) DOWN 1 R22 2 ELECTRIC (11) A11 R51 1ST AP (57) DOWN 1 R22 3 ELECTRIC (12) A12 R51 1ST AP (58) DOWN 1 R22 3 ELECTRIC (13) A13 R51 1ST AP (59) DOWN 1 R22 3 ELECTRIC (14) A14 R51 1ST AP (60) DOWN 1 R22 4 ELECTRIC (15) A15 R51 1ST AP (61) DOWN 1 R22 4 ELECTRIC (16) A16 R51 1 ELECTRIC (62) DOWN 1 R22 4 ELECTRIC (17) A17 R51 1 ELECTRIC (63) DOWN 1 R22 4A ELECTRIC (18) A18 R51 1 ELECTRIC (64) DOWN 1 R22 4A ELECTRIC (19) A19 R51 1 ELECTRIC (65) DOWN 1 R22 4A ELECTRIC (20) A20 R51 2 ELECTRIC (71) DOWN 2 R80 1 ELECTRIC (21) A21 R51 2 ELECTRIC (72) DOWN 2 R80 1 ELECTRIC (22) A22 R51 2 ELECTRIC (73) DOWN 2 R80 1 ELECTRIC (23) A23 R51 2 ELECTRIC (74) DOWN 2 R80 2 ELECTRIC (24) A24 R51 3 ELECTRIC (75) DOWN 2 R80 2 ELECTRIC (25) A25 R51 3 ELECTRIC (76) DOWN 2 R80 2 ELECTRIC (26) A26 R51 3 ELECTRIC (77) DOWN 2 R80 3 ELECTRIC (27) A27 R51 3 ELECTRIC (78) DOWN 2 R80 3 ELECTRIC (28) A28 R51 4 ELECTRIC (79) DOWN 2 R80 3 ELECTRIC (29) A29 R51 4 ELECTRIC (80) DOWN 2 R80 4 ELECTRIC (30) A30 R51 4 ELECTRIC (81) DOWN 2 R80 4 ELECTRIC (31) A31 R51 4 ELECTRIC (82) DOWN 2 R80 4 ELECTRIC (41) APRON R51 1ST AP (83) DOWN 2 R80 4A ELECTRIC (42) APRON .. -

USCIS Employment Authorization Documents
USCIS Employment Authorization Documents March 19, 2018 Fiscal Year 2017 Report to Congress U.S. Citizenship and Immigration Services Message from U.S. Citizenship and Immigration Services March 19, 2018 I am pleased to present the following report, “USCIS Employment Authorization Documents,” which has been prepared by U.S. Citizenship and Immigration Services (USCIS). This report was compiled pursuant to language set forth in Senate Report 114-264 accompanying the Fiscal Year (FY) 2017 Department of Homeland Security Appropriations Act (P.L. 115-31). Pursuant to congressional requirements, this report is being provided to the following Members of Congress: The Honorable John R. Carter Chairman, House Appropriations Subcommittee on Homeland Security The Honorable Lucille Roybal-Allard Ranking Member, House Appropriations Subcommittee on Homeland Security The Honorable John Boozman Chairman, Senate Appropriations Subcommittee on Homeland Security The Honorable Jon Tester Ranking Member, Senate Appropriations Subcommittee on Homeland Security I am pleased to respond to any questions you may have. Please do not hesitate to contact me at (202) 272-1000 or the Department’s Acting Chief Financial Officer, Stacy Marcott, at (202) 447-5751. Sincerely, L. Francis Cissna Director U.S. Citizenship and Immigration Services i Executive Summary This report provides the information requested by the Senate Appropriations Committee regarding the number of employment authorization documents (EAD) issued annually from FY 2012 through FY 2015, the validity period of those EADs, and the policies governing validity periods of EADs. As requested, the report provides details on the number and type of EAD approvals by USCIS. From FYs 2012–2015, USCIS approved more than 6 million EADs in multiple categories. -

Identifying Effective Features and Classifiers for Short Term Rainfall Forecast Using Rough Sets Maximum Frequency Weighted Feature Reduction Technique
CIT. Journal of Computing and Information Technology, Vol. 24, No. 2, June 2016, 181–194 181 doi: 10.20532/cit.2016.1002715 Identifying Effective Features and Classifiers for Short Term Rainfall Forecast Using Rough Sets Maximum Frequency Weighted Feature Reduction Technique Sudha Mohankumar1 and Valarmathi Balasubramanian2 1Information Technology Department, School of Information Technology and Engineering, VIT University, Vellore, India 2Software and Systems Engineering Department, School of Information Technology and Engineering, VIT University, Vellore, India Precise rainfall forecasting is a common challenge Information systems → Information retrieval → Re- across the globe in meteorological predictions. As trieval tasks and goals → Clustering and classification; rainfall forecasting involves rather complex dy- Applied computing → Operations research → Fore- namic parameters, an increasing demand for novel casting approaches to improve the forecasting accuracy has heightened. Recently, Rough Set Theory (RST) has at- Keywords: rainfall prediction, rough set, maximum tracted a wide variety of scientific applications and is frequency, optimal reduct, core features and accuracy extensively adopted in decision support systems. Al- though there are several weather prediction techniques in the existing literature, identifying significant input for modelling effective rainfall prediction is not ad- dressed in the present mechanisms. Therefore, this in- 1. Introduction vestigation has examined the feasibility of using rough set based feature selection and data mining methods, Rainfall forecast serves as an important di- namely Naïve Bayes (NB), Bayesian Logistic Re- saster prevention tool. Agricultural yields and gression (BLR), Multi-Layer Perceptron (MLP), J48, agriculture based industrial development more Classification and Regression Tree (CART), Random often than not, rely on natural water resources Forest (RF), and Support Vector Machine (SVM), to forecast rainfall. -

Specification for Welding Shielding Gases
ANSI/AWS A5.32/A5.32M-97 An American National Standard Specification for Welding Shielding Gases Key Words— Argon, carbon dioxide, helium, ANSI/AWS A5.32/A5.32M-97 hydrogen, nitrogen, oxygen, shielding An American National Standard gases, welding gases Approved by American National Standards Institute December 8, 1997 Specification for Welding Shielding Gases Prepared by AWS Committee on Filler Metals Under the Direction of AWS Technical Activities Committee Approved by AWS Board of Directors Abstract This specification for welding shielding gases specifies minimum requirements for the composition and purity of the most popular single-component shielding gases. Classification designators for both single and multicomponent gases are introduced. Other topics include testing procedures, package marking, and general application guidelines. This specification makes use of both U.S. Customary Units and the International System of Units (SI). Since these are not equivalent, each system must be used independently of the other. 550 N.W. LeJeune Road, Miami, Florida 33126 Table of Contents Page No. Personnel.................................................................................................................................................................... iii Foreword ................................................................................................................................................................... v List of Tables ..............................................................................................................................................................vii