Efficient Identification of a Novel Cancer/Testis Antigen For
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplemental Figures
Supplemental Figures Development of a new macrophage-specific TRAP mouse (MacTRAP) and definition of the renal macrophage translational signature Andreas Hofmeister, Maximilian C. Thomassen, Sabrina Markert, André Marquardt, Mathieu Preußner, Martin Rußwurm, Ralph Schermuly, Ulrich Steinhoff, Hermann-Josef Gröne, Joachim Hoyer, Benjamin D. Humphreys, Ivica Grgic Correspondence: Ivica Grgic MD, Klinikum der Philips-Universität Marburg, Baldingerstrasse 1, 35043 Marburg. Phone: +4964215861736, email: [email protected] Sup. Fig. S1 Δ6.7fmsGFP-L10a plasmid (pGL2 backbone) Sup. Fig. S1: Plasmid map of the engineered c-fms-eGFP-L10a expression vector. Mlu1/Sal1 digestion was used for linearization and extraction of the transgene. Sup. Fig. S2 A Neutrophils Monocytes Lymphocytes B Monocytes Monocytes Neutrophils Lymphocytes H - TRAP Ly6g Count FSC CD115 Mac GFP GFP GFP GFP H - Ly6g Count FSC CD115 Wild Wild type GFP GFP GFP GFP Sup. Fig. S2: FACS analysis detects eGFP-L10a signals in monocytes but not in neutrophils or lymphocytes isolated from peripheral blood of MacTRAP mice. (A) Gating strategy to define monocyte, neutrophil and lymphocyte populations. Only single cells contributed to the analysis. (B) GFP-fluorescence was specifically detected in CD115+ monocytes, but not in Ly6g+ neutrophils or lymphocytes of MacTRAP mice. Blood samples from wild-type mice served as negative controls. Representative plots are shown, n=6. Sup. Fig. S3 A eGFP-L10a Ly6g merge+DAPI kidney 7d UUO B eGFP-L10a Ly6g merge+DAPI incision skin Tail Sup. Fig. S3: Immunostaining for mature neutrophils in fibrotic kidney tissue and tail skin biopsies from MacTRAP mice. (A) Only a very small fraction of Ly6g+ neutrophils is positive for eGFP-L10a in fibrotic kidneys after 7d UUO (0.42% ± 0.29%; 468 cells counted, n=3). -

The Genetic Program of Pancreatic Beta-Cell Replication in Vivo
Page 1 of 65 Diabetes The genetic program of pancreatic beta-cell replication in vivo Agnes Klochendler1, Inbal Caspi2, Noa Corem1, Maya Moran3, Oriel Friedlich1, Sharona Elgavish4, Yuval Nevo4, Aharon Helman1, Benjamin Glaser5, Amir Eden3, Shalev Itzkovitz2, Yuval Dor1,* 1Department of Developmental Biology and Cancer Research, The Institute for Medical Research Israel-Canada, The Hebrew University-Hadassah Medical School, Jerusalem 91120, Israel 2Department of Molecular Cell Biology, Weizmann Institute of Science, Rehovot, Israel. 3Department of Cell and Developmental Biology, The Silberman Institute of Life Sciences, The Hebrew University of Jerusalem, Jerusalem 91904, Israel 4Info-CORE, Bioinformatics Unit of the I-CORE Computation Center, The Hebrew University and Hadassah, The Institute for Medical Research Israel- Canada, The Hebrew University-Hadassah Medical School, Jerusalem 91120, Israel 5Endocrinology and Metabolism Service, Department of Internal Medicine, Hadassah-Hebrew University Medical Center, Jerusalem 91120, Israel *Correspondence: [email protected] Running title: The genetic program of pancreatic β-cell replication 1 Diabetes Publish Ahead of Print, published online March 18, 2016 Diabetes Page 2 of 65 Abstract The molecular program underlying infrequent replication of pancreatic beta- cells remains largely inaccessible. Using transgenic mice expressing GFP in cycling cells we sorted live, replicating beta-cells and determined their transcriptome. Replicating beta-cells upregulate hundreds of proliferation- related genes, along with many novel putative cell cycle components. Strikingly, genes involved in beta-cell functions, namely glucose sensing and insulin secretion were repressed. Further studies using single molecule RNA in situ hybridization revealed that in fact, replicating beta-cells double the amount of RNA for most genes, but this upregulation excludes genes involved in beta-cell function. -

(12) Patent Application Publication (10) Pub. No.: US 2009/0113561 A1 Von Melchner Et Al
US 2009.0113561A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2009/0113561 A1 Von Melchner et al. (43) Pub. Date: Apr. 30, 2009 (54) GENE TRAP CASSETTES FOR RANDOM (30) Foreign Application Priority Data AND TARGETED CONDITIONAL GENE NACTIVATION Nov. 26, 2004 (EP) .................................. O4O281941 Apr. 18, 2005 (EP) .................................. O5103092.2 (75) Inventors: Harald Von Melchner, Publication Classification Kronberg/Taunus (DE); Frank (51) Int. Cl Schnutgen, Alzenau (DE); AOIK 67/027 (2006.01) Particia Ruiz, Berlin (DE): Silke CI2N 15/87 (2006.01) De-Zolt, Rodenbach (DE); Thomas CI2O I/68 (2006.01) Floss, Oberappersdorf (DE); Jens CI2N 5/06 (2006.01) Hansen, Kirchheim (DE) (52) U.S. Cl. .............. 800/3:536/23.1; 435/325; 800/13; 435/455: 800/25; 435/6:435/463 Correspondence Address: (57) ABSTRACT NORRIS, MCLAUGHILIN & MARCUS, PA 875 THIRDAVENUE, 18TH FLOOR A new type of gene trap cassette, which can induce condi NEW YORK, NY 10022 (US) tional mutations, relies on directional site-specific recombi nation systems, which can repair and re-induce gene trap (73) Assignee: FRANKGEN mutations when activated in Succession. After the gene trap cassettes are inserted into the genome of the target organism, BIOTECHNOLOGIE AG, mutations can be activated at a particular time and place in Kronberg (DE) Somatic cells. The gene trap cassettes also create multipur pose alleles amendable to a wide range of post-insertional (21) Appl. No.: 11/720,231 modifications. Such gene trap cassettes can be used to muta tionally inactivate all cellular genes temporally and/or spa (22) PCT Filed: Nov. -

Identification of Genomic Targets of Krüppel-Like Factor 9 in Mouse Hippocampal
Identification of Genomic Targets of Krüppel-like Factor 9 in Mouse Hippocampal Neurons: Evidence for a role in modulating peripheral circadian clocks by Joseph R. Knoedler A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Neuroscience) in the University of Michigan 2016 Doctoral Committee: Professor Robert J. Denver, Chair Professor Daniel Goldman Professor Diane Robins Professor Audrey Seasholtz Associate Professor Bing Ye ©Joseph R. Knoedler All Rights Reserved 2016 To my parents, who never once questioned my decision to become the other kind of doctor, And to Lucy, who has pushed me to be a better person from day one. ii Acknowledgements I have a huge number of people to thank for having made it to this point, so in no particular order: -I would like to thank my adviser, Dr. Robert J. Denver, for his guidance, encouragement, and patience over the last seven years; his mentorship has been indispensable for my growth as a scientist -I would also like to thank my committee members, Drs. Audrey Seasholtz, Dan Goldman, Diane Robins and Bing Ye, for their constructive feedback and their willingness to meet in a frequently cold, windowless room across campus from where they work -I am hugely indebted to Pia Bagamasbad and Yasuhiro Kyono for teaching me almost everything I know about molecular biology and bioinformatics, and to Arasakumar Subramani for his tireless work during the home stretch to my dissertation -I am grateful for the Neuroscience Program leadership and staff, in particular -

Induction of Therapeutic Tissue Tolerance Foxp3 Expression Is
Downloaded from http://www.jimmunol.org/ by guest on October 2, 2021 is online at: average * The Journal of Immunology , 13 of which you can access for free at: 2012; 189:3947-3956; Prepublished online 17 from submission to initial decision 4 weeks from acceptance to publication September 2012; doi: 10.4049/jimmunol.1200449 http://www.jimmunol.org/content/189/8/3947 Foxp3 Expression Is Required for the Induction of Therapeutic Tissue Tolerance Frederico S. Regateiro, Ye Chen, Adrian R. Kendal, Robert Hilbrands, Elizabeth Adams, Stephen P. Cobbold, Jianbo Ma, Kristian G. Andersen, Alexander G. Betz, Mindy Zhang, Shruti Madhiwalla, Bruce Roberts, Herman Waldmann, Kathleen F. Nolan and Duncan Howie J Immunol cites 35 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription http://www.jimmunol.org/content/suppl/2012/09/17/jimmunol.120044 9.DC1 This article http://www.jimmunol.org/content/189/8/3947.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2012 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of October 2, 2021. -

Plasticity and the Impact of Increasing Temperature on a Tropical Ectotherm
Georgia Southern University Digital Commons@Georgia Southern Electronic Theses and Dissertations Graduate Studies, Jack N. Averitt College of Spring 2020 Plasticity and the Impact of Increasing Temperature on a Tropical Ectotherm Adam A. Rosso Follow this and additional works at: https://digitalcommons.georgiasouthern.edu/etd Part of the Bioinformatics Commons, Comparative and Evolutionary Physiology Commons, Evolution Commons, Integrative Biology Commons, Other Ecology and Evolutionary Biology Commons, and the Other Genetics and Genomics Commons Recommended Citation Rosso, Adam A., "Plasticity and the Impact of Increasing Temperature on a Tropical Ectotherm" (2020). Electronic Theses and Dissertations. 2069. https://digitalcommons.georgiasouthern.edu/etd/2069 This thesis (open access) is brought to you for free and open access by the Graduate Studies, Jack N. Averitt College of at Digital Commons@Georgia Southern. It has been accepted for inclusion in Electronic Theses and Dissertations by an authorized administrator of Digital Commons@Georgia Southern. For more information, please contact [email protected]. PLASTICITY AND THE IMPAPCT OF INCREASING TEMPERATURE ON A TROPICAL ECTOTHERM by ADAM A. ROSSO (Under the direction of Christian L. Cox) ABSTRACT Organisms may respond to climate change through behavior, genetic adaptation, and/or phenotypic plasticity. Tropical ectotherms are thought to be especially vulnerable to climate change because most have a narrow range of thermal tolerance while living close to their upper thermal tolerance limits. Additionally, many tropical species live in closed-canopy forests, which provide homogenous thermal landscapes that prevent behavioral compensation for stressfully warm temperatures. Finally, tropical ectotherms are thought to have decreased capacity for phenotypic plasticity because they have evolved in thermally stable environments. -
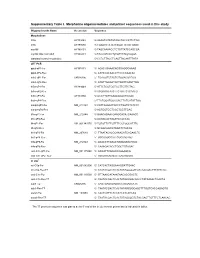
Supplementary Table I. Morpholino Oligonucleotides and Primer Sequences Used in This Study
Supplementary Table I. Morpholino oligonucleotides and primer sequences used in this study Oligonucleotide Name Accession Sequence Morpholinos tlr5a AY389449 5'-AAAGTGTATGTAGCTGCCATTCTGG tlr5b AY389450 5'-TGAATGTATATCCCATTCTGTGAGC myd88 AY388401 5'-TAGCAAAACCTCTGTTATCCAGCGA myd88 5bp mismatch AY388401 5'-TAcCAtAACCTgTGTTATCgAGgGA standard control morpholino 5'-CCTCTTACCTCAGTTACAATTTATA qRT-PCR ppial-qP1-Fw AY391451 5’- ACACTGAAACACGGAGGCAAAG ppial-qP2-Rev 5’- CATCCACAACCTTCCCGAACAC irak3-qP1-Fw CK026195 5’- TGAGGTCTACTGTGGACGATGG irak3-qP2-Rev 5’- ATGTTAGGATGCTGGTTGAGTTGG tlr5a-qP1-Fw AY389449 5’-ATTCTGGTGGTGCTTGTTGTAG tlr5a-qP2-Rev 5’-ACGAGGTAACTTCTGTTCTCAATG tlr5b-qP3-Fw AY389450 5’-GCGTTGTTGAAGAGGCTGGAC tlr5b-qP4-Rev 5’-TTCTGGATGGCCACTTCTCATATTGG mmp9-qP3-Fw NM_213123 5’-CATTAAAGATGCCCTGATGTATCCC mmp9-qP4-Rev 5’-AGTGGTGGTCCGTGGTTGAG il1b-qP1-Fw NM_212844 5’-GAACAGAATGAAGCACATCAAACC il1b-qP2-Rev 5’-ACGGCACTGAATCCACCAC il8-qP1-Fw XM_001342570 5’-TGTGTTATTGTTTTCCTGGCATTTC il8-qP2-Rev 5’-GCGACAGCGTGGATCTACAG ifn1-qP3-Fw NM_207640 5’- TTAATACACGCAAAGATGAGAACTC ifn1-qP4-Rev 5’- GCCAAGCCATTCGCAAGTAG tnfa-qP5-Fw NM_212829 5’- AGACCTTAGACTGGAGAGATGAC tnfa-qP6-Rev 5’- CAAAGACACCTGGCTGTAGAC cxcl-C1c-qP1-Fw NM_001115060 5’- GGCATTCACACCCAAAGCG cxcl-C1c-qP2_Rev 5’- GCGAGCACGATTCACGAGAG * In situ ccl-C5a-Fw NM_001082906 5’- CATCACTAGGAAAGGATTGAAC ccl-C5a-Rev-T7 5’- TAATACGACTCACTATAGGGGATGTCAAAGACTTTATTCAC cxcl-C1c-Fw NM_001115060 5’- GTTAAACATAAATAACACCGACTC cxcl-C1c-Rev-T7 5’- TAATACGACTCACTATAGGGACACCCTATAAAACTGAGTA irak3-Fw CK026195 5’- CAGTGAGAGAGGCATGAAACATC -

List of Genes Deregulated in HDAC1 KO Vs WT
Yamaguchi_Suppl. Table 1 List of genes deregulated in HDAC1 KO vs WT Probe Set name fold change 1423327_at RIKEN cDNA 4930517K11 gene 201,6 1426231_at vitrin 33,4 1416368_at glutathione S-transferase, alpha 4 23,2 1435436_at Transcribed locus 20,1 1456379_x_at delta/notch-like EGF-related receptor 15,2 1418175_at vitamin D receptor 14,7 1455642_a_at tetraspanin 17 14,1 1436448_a_at prostaglandin-endoperoxide synthase 1 13,7 1423414_at prostaglandin-endoperoxide synthase 1 12,3 1416710_at transmembrane protein 35 11,8 1454780_at UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 4 10,9 1439231_at gb:BG228852 /DB_XREF=gi:12716356 /DB_XREF=ux62e05.x1 /CLONE=IMAGE:3514856 /FEA=EST /CNT=9 /TID=Mm.133155.1 /TIER=Stack10,3 /STK=8 /UG=Mm.133155 /UG_TITLE=ESTs 1416072_at CD34 antigen 10,2 1433529_at RIKEN cDNA E430002G05 gene 9,2 1416203_at aquaporin 1 8,6 1418949_at growth differentiation factor 15 8,3 1435955_at sialic acid binding Ig-like lectin 10 8,2 1416326_at cysteine-rich protein 1 (intestinal) 8,2 1448700_at G0/G1 switch gene 2 8,1 1416271_at PERP, TP53 apoptosis effector 8,0 1418176_at vitamin D receptor 7,8 1439789_at gb:BQ177189 /DB_XREF=gi:20352681 /DB_XREF=UI-M-DJ2-bwa-j-16-0-UI.s1 /CLONE=UI-M-DJ2-bwa-j-16-0-UI /FEA=EST /CNT=16 /TID=7,4 Mm.34073.1 /TIER=ConsEnd /STK=6 /UG=Mm.34073 /UG_TITLE=ESTs 1423627_at NAD(P)H dehydrogenase, quinone 1 7,3 1442174_at tetraspanin 18 7,3 1435945_a_at potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 6,9 1439362_at Transcribed -

Datasheet Blank Template
SAN TA C RUZ BI OTEC HNOL OG Y, INC . Nup43 (6L4): sc-134406 BACKGROUND APPLICATIONS The nuclear pore complex (NPC) mediates bidirectional macromolecular traffic Nup43 (6L4) is recommended for detection of Nup43 of human origin by between the nucleus and cytoplasm in eukaryotic cells and is comprised of Western Blotting (starting dilution 1:200, dilution range 1:100-1:1000), more than 100 different subunits. Many of the subunits belong to a family immunoprecipitation [1-2 µg per 100-500 µg of total protein (1 ml of cell called nucleoporins (Nups), which are characterized by the presence of O- lysate)] and solid phase ELISA (starting dilution 1:30, dilution range 1:30- linked-N-acetylglucosamine moieties and a distinctive pentapeptide repeat 1:3000). (XFXFG). Nup43 (nucleoporin 43 kDa) is a 380 amino acid protein that contains Suitable for use as control antibody for Nup43 siRNA (h): sc-95534, Nup43 six WD domains and exists as a component of the Nup107-160 subcomplex shRNA Plasmid (h): sc-95534-SH and Nup43 shRNA (h) Lentiviral Particles: of the nuclear pore complex (NPC). The Nup107-160 subcomplex is required sc-95534-V. for the assembly of a functional NPC as well as for normal kinetochore micro - tubule attachment, mitotic progression and chromosome segregation. Mem- Molecular Weight of Nup43: 43 kDa. bers of the Nup107-160 subcomplex include NUP160, NUP133, NUP107, Positive Controls: human Nup43 transfected 293T whole cell lysate. NUP98, NUP85, NUP37, SEH1 and SEC13. RECOMMENDED SUPPORT REAGENTS REFERENCES To ensure optimal results, the following support reagents are recommended: 1. Schlenstedt, G. -

(12) Patent Application Publication (10) Pub. No.: US 2009/0163434 A1 BADER Et Al
US 200901 63434A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2009/0163434 A1 BADER et al. (43) Pub. Date: Jun. 25, 2009 (54) MIR-20 REGULATED GENES AND Related U.S. Application Data PATHWAYS AS TARGETS FOR THERAPEUTIC INTERVENTION (60) Provisional application No. 60/915,026, filed on Apr. 30, 2007. (76) Inventors: Andreas G. BADER, Austin, TX Publication Classification (US); Mike BYROM, Austin, TX (US); Charles D. JOHNSON, (51) Int. Cl. Austin, TX (US); David BROWN, A6IR 48/00 (2006.01) Austin, TX (US) CI2O I/68 (2006.01) A6IP 43/00 (2006.01) Correspondence Address: (52) U.S. Cl. ................................ 514/44; 435/6; 977/773 Fulbright & Jaworski L.L.P. 600 Congress Avenue, Suite 2400 (57) ABSTRACT Austin, TX 78701 (US) The present invention concerns methods and compositions for identifying genes or genetic pathways modulated by miR (21) Appl. No.: 12/112,291 20a, using miR-20a to modulate a gene or gene pathway, using this profile in assessing the condition of a patient and/or (22) Filed: Apr. 30, 2008 treating the patient with an appropriate miRNA. US 2009/0163434 A1 Jun. 25, 2009 MR-2O REGULATED GENES AND cers (reviewed in Esquela-Kerscher and Slack, 2006; Calin PATHWAYS AS TARGETS FOR and Croce, 2006). Differential expression of almost all miR THERAPEUTIC INTERVENTION NAS across numerous cancer types has been observed (Lu et al., 2005). Most such studies link miRNAs to cancer only by indirect evidence. However. He et al. (2005a) has provided 0001. This application claims priority to U.S. Provisional more direct evidence that miRNAs may contribute directly to Patent application Ser. -

Supplemental Figure 1
A BCD6 35 80 1.0 3 Thr Ala 0.4 Thr Ala 30 0.8 60 4 0.3 2 25 0.6 40 20 0.2 0.4 1 2 20 intake(g)Food Body weight(g) 15 0.1 0.2 Serum TSH (ng/ml) T4 plasmaT4 levels (ng/ml) plasma levelsT3 (ng/ml) 1/V 10 0.0 0 0.0 0 5 10 15 Thr Ala Thr Ala Thr Ala -1.0 -0.5 0.5 1.0 1.5 2.0 Thr Ala Thr Ala Age (weeks) 1/[T4] (nM) dark light (fmols/min/mg ptn) (fmols/min/mg -1 Ala Thr IKMN O Femur 16.5 0.55 1100 0.24 E F 16.0 1050 0.22 G H 0.50 Thr92‐D2 15.5 1000 0.20 Ala92‐D2 0.45 15.0 950 0.18 (mgHAcm-3) Femur volume 0.40 14.5 900 0.16 Cortical thickness Femur length (mm) length Femur Bone MineralDensity 14.0 0.35 850 0.14 Thr Ala Thr Ala Thr Ala Thr Ala JLPQR Vertebrae 4.6 5.5 0.070 0.35 4.4 5.0 0.065 0.30 **P<0.01 0.060 4.2 *** 4.5 0.055 0.25 4.0 4.0 0.050 0.20 3.8 3.5 Trabecular number 0.045 Trabecular spacing Trabecular thickness Vertebral height (mm) height Vertebral 3.6 3.0 0.040 0.15 Thr Ala Thr Ala Thr Ala Thr Ala Figure S1: Additional phenotype of the Ala92‐Dio2 mouse. -
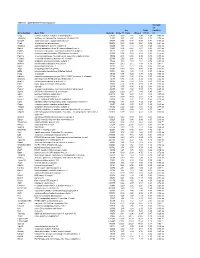
Table S-1 Mpkccd Transcriptome
Table S1. mpkCCD Cell Transcriptomes. Correlati on Ori. Ratio Coefficie Gene Symbol Gene Title GeneID Clone 11 Clone Clone 2 (11/2) nt Copg coatomer protein complex, subunit gamma 54161 1.99 1.81 2.48 0.80 -0.49 ns Atp6v0d1 ATPase, H+ transporting, lysosomal V0 subunit D1 11972 3.37 2.87 3.00 1.12 0.74 ns Golga7 golgi autoantigen, golgin subfamily a, 7 57437 3.03 3.40 3.58 0.84 -0.34 ns Psph phosphoserine phosphatase 100678 0.38 0.66 0.64 0.59 -0.61 ns Trappc4 trafficking protein particle complex 4 60409 1.17 1.12 1.08 1.08 0.64 ns Dpm2 dolichol-phosphate (beta-D) mannosyltransferase 2 13481 0.74 0.87 0.81 0.91 -0.31 ns Psmb5 proteasome (prosome, macropain) subunit, beta type 5 19173 2.92 3.00 2.92 0.99 0.32 ns Dhrs1 dehydrogenase/reductase (SDR family) member 1 52585 0.74 0.68 0.88 0.83 -0.18 ns Ppm1a protein phosphatase 1A, magnesium dependent, alpha isoform 19042 2.43 2.60 2.96 0.82 -0.53 ns Psenen presenilin enhancer 2 homolog (C. elegans) 66340 2.52 2.88 3.80 0.66 -0.77 ns Anapc1 anaphase promoting complex subunit 1 17222 1.16 1.33 1.61 0.72 -0.33 ns Mrpl43 mitochondrial ribosomal protein L43 94067 2.15 2.15 1.84 1.16 0.91 * Nmt1 N-myristoyltransferase 1 18107 3.21 2.71 3.36 0.95 0.31 ns Atg5 autophagy-related 5 (yeast) 11793 0.70 0.73 0.66 1.05 -0.23 ns Mtif2 mitochondrial translational initiation factor 2 76784 1.16 1.29 1.19 0.97 -0.36 ns Psap prosaposin 19156 8.07 7.25 8.37 0.96 0.02 ns Ube2g1 ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, C.