(12) Patent Application Publication (10) Pub. No.: US 2009/0163434 A1 BADER Et Al
Total Page:16
File Type:pdf, Size:1020Kb

Load more
Recommended publications
-

The Switch from Fermentation to Respiration in Saccharomyces Cerevisiae Is Regulated by the Ert1 Transcriptional Activator/Repressor
INVESTIGATION The Switch from Fermentation to Respiration in Saccharomyces cerevisiae Is Regulated by the Ert1 Transcriptional Activator/Repressor Najla Gasmi,* Pierre-Etienne Jacques,† Natalia Klimova,† Xiao Guo,§ Alessandra Ricciardi,§ François Robert,†,** and Bernard Turcotte*,‡,§,1 ‡Department of Medicine, *Department of Biochemistry, and §Department of Microbiology and Immunology, McGill University Health Centre, McGill University, Montreal, QC, Canada H3A 1A1, †Institut de recherches cliniques de Montréal, Montréal, QC, Canada H2W 1R7, and **Département de Médecine, Faculté de Médecine, Université de Montréal, QC, Canada H3C 3J7 ABSTRACT In the yeast Saccharomyces cerevisiae, fermentation is the major pathway for energy production, even under aerobic conditions. However, when glucose becomes scarce, ethanol produced during fermentation is used as a carbon source, requiring a shift to respiration. This adaptation results in massive reprogramming of gene expression. Increased expression of genes for gluconeogenesis and the glyoxylate cycle is observed upon a shift to ethanol and, conversely, expression of some fermentation genes is reduced. The zinc cluster proteins Cat8, Sip4, and Rds2, as well as Adr1, have been shown to mediate this reprogramming of gene expression. In this study, we have characterized the gene YBR239C encoding a putative zinc cluster protein and it was named ERT1 (ethanol regulated transcription factor 1). ChIP-chip analysis showed that Ert1 binds to a limited number of targets in the presence of glucose. The strongest enrichment was observed at the promoter of PCK1 encoding an important gluconeogenic enzyme. With ethanol as the carbon source, enrichment was observed with many additional genes involved in gluconeogenesis and mitochondrial function. Use of lacZ reporters and quantitative RT-PCR analyses demonstrated that Ert1 regulates expression of its target genes in a manner that is highly redundant with other regulators of gluconeogenesis. -

Supplemental Material
Supplemental Table B ARGs in alphabetical order Symbol Title 3 months 6 months 9 months 12 months 23 months ANOVA Direction Category 38597 septin 2 1557 ± 44 1555 ± 44 1579 ± 56 1655 ± 26 1691 ± 31 0.05219 up Intermediate 0610031j06rik kidney predominant protein NCU-G1 491 ± 6 504 ± 14 503 ± 11 527 ± 13 534 ± 12 0.04747 up Early Adult 1G5 vesicle-associated calmodulin-binding protein 662 ± 23 675 ± 17 629 ± 16 617 ± 20 583 ± 26 0.03129 down Intermediate A2m alpha-2-macroglobulin 262 ± 7 272 ± 8 244 ± 6 290 ± 7 353 ± 16 0.00000 up Midlife Aadat aminoadipate aminotransferase (synonym Kat2) 180 ± 5 201 ± 12 223 ± 7 244 ± 14 275 ± 7 0.00000 up Early Adult Abca2 ATP-binding cassette, sub-family A (ABC1), member 2 958 ± 28 1052 ± 58 1086 ± 36 1071 ± 44 1141 ± 41 0.05371 up Early Adult Abcb1a ATP-binding cassette, sub-family B (MDR/TAP), member 1A 136 ± 8 147 ± 6 147 ± 13 155 ± 9 185 ± 13 0.01272 up Midlife Acadl acetyl-Coenzyme A dehydrogenase, long-chain 423 ± 7 456 ± 11 478 ± 14 486 ± 13 512 ± 11 0.00003 up Early Adult Acadvl acyl-Coenzyme A dehydrogenase, very long chain 426 ± 14 414 ± 10 404 ± 13 411 ± 15 461 ± 10 0.01017 up Late Accn1 amiloride-sensitive cation channel 1, neuronal (degenerin) 242 ± 10 250 ± 9 237 ± 11 247 ± 14 212 ± 8 0.04972 down Late Actb actin, beta 12965 ± 310 13382 ± 170 13145 ± 273 13739 ± 303 14187 ± 269 0.01195 up Midlife Acvrinp1 activin receptor interacting protein 1 304 ± 18 285 ± 21 274 ± 13 297 ± 21 341 ± 14 0.03610 up Late Adk adenosine kinase 1828 ± 43 1920 ± 38 1922 ± 22 2048 ± 30 1949 ± 44 0.00797 up Early -

Original Article Dynamics of Lamins B and A/C and Nucleoporin Nup160 During Meiotic Maturation in Mouse Oocytes
Original Article Dynamics of Lamins B and A/C and Nucleoporin Nup160 during Meiotic Maturation in Mouse Oocytes (oocytes / meiosis / meiotic spindle / nuclear lamina / Nup107-160 / nuclear pore complex) V. NIKOLOVA, S. DELIMITREVA, I. CHAKAROVA, R. ZHIVKOVA, V. HADZHINESHEVA, M. MARKOVA Department of Biology, Medical Faculty, Medical University of Sofia, Bulgaria Abstract. This study was aimed at elucidating the plex reorganization of the cytoskeleton and nuclear en- fate of three important nuclear envelope components velope (Delimitreva et al., 2012). Although the early – lamins B and A/C and nucleoporin Nup160, during meiotic stages have been relatively well studied, the meiotic maturation of mouse oocytes. These proteins events of final steps of oocyte meiosis (from meiotic re- were localized by epifluorescence and confocal mi- sumption in late prophase I until metaphase II) are still croscopy using specific antibodies in oocytes at dif- poorly understood. The oocyte nucleus in late prophase ferent stages from prophase I (germinal vesicle) to I, traditionally called GV (germinal vesicle), becomes metaphase II. In immature germinal vesicle oocytes, competent to resume meiosis upon accumulation of all three proteins were detected at the nuclear pe- pericentriolar heterochromatin called karyosphere, sur- riphery. In metaphase I and metaphase II, lamin B rounded nucleolus or rimmed nucleolus (Can et al., co-localized with the meiotic spindle, lamin A/C was 2003; De la Fuente et al., 2004; Tan et al., 2009). Then, found in a diffuse halo surrounding the spindle and the nucleus disaggregates in the so-called germinal ve- to a lesser degree throughout the cytoplasm, and sicle breakdown (GVBD) stage. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

New Targets of Urocortin-Mediated Cardioprotection
69 New targets of urocortin-mediated cardioprotection Sea´n P Barry1, Kevin M Lawrence4, James McCormick1, Surinder M Soond5, Mike Hubank2, Simon Eaton3, Ahila Sivarajah6, Tiziano M Scarabelli7, Richard A Knight1, Christoph Thiemermann6, David S Latchman1, Paul A Townsend8 and Anastasis Stephanou1 1Medical Molecular Biology Unit, 2Department of Molecular Haematology and 3Department of Surgery, Institute of Child Health, University College London, 30 Guilford Street, London, WC1N 1EH, UK 4Department of Cellular Pathology, St George’s, University of London, Cranmer Terrace, Tooting, London, SW17 0RE, UK 5School of Biological Sciences, University of East Anglia, Norwich, NR4 7TJ, UK 6St Bartholomew’s and The Royal London School of Medicine and Dentistry, William Harvey Research Institute, Centre for Translational Medicine and Therapeutics, Queen Mary University of London, London, EC1M 7BQ, UK 7Center for Heart and Vessel Preclinical Studies, St John Hospital and Medical Center, Wayne State University School of Medicine, 22201 Moross Road, Detroit, Michigan 48336, USA 8Human Genetics Division, MP808, Southampton General Hospital, University of Southampton, Southampton SO16 6YD, UK (Correspondence should be addressed to S P Barry; Email: [email protected]) Abstract The urocortin (UCN) hormones UCN1 and UCN2 have been shown previously to confer significant protection against myocardial ischaemia/reperfusion (I/R) injury; however, the molecular mechanisms underlying their action are poorly understood. To further define the transcriptional effect of UCNs that underpins their cardioprotective activity, a microarray analysis was carried out using an in vivo rat coronary occlusion model of I/R injury. Infusion of UCN1 or UCN2 before the onset of reperfusion resulted in the differential regulation of 66 and 141 genes respectively, the majority of which have not been described previously. -

Antigen-Specific Memory CD4 T Cells Coordinated Changes in DNA
Downloaded from http://www.jimmunol.org/ by guest on September 24, 2021 is online at: average * The Journal of Immunology The Journal of Immunology published online 18 March 2013 from submission to initial decision 4 weeks from acceptance to publication http://www.jimmunol.org/content/early/2013/03/17/jimmun ol.1202267 Coordinated Changes in DNA Methylation in Antigen-Specific Memory CD4 T Cells Shin-ichi Hashimoto, Katsumi Ogoshi, Atsushi Sasaki, Jun Abe, Wei Qu, Yoichiro Nakatani, Budrul Ahsan, Kenshiro Oshima, Francis H. W. Shand, Akio Ametani, Yutaka Suzuki, Shuichi Kaneko, Takashi Wada, Masahira Hattori, Sumio Sugano, Shinichi Morishita and Kouji Matsushima J Immunol Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Author Choice option Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Freely available online through http://www.jimmunol.org/content/suppl/2013/03/18/jimmunol.120226 7.DC1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material Permissions Email Alerts Subscription Author Choice Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 24, 2021. Published March 18, 2013, doi:10.4049/jimmunol.1202267 The Journal of Immunology Coordinated Changes in DNA Methylation in Antigen-Specific Memory CD4 T Cells Shin-ichi Hashimoto,*,†,‡ Katsumi Ogoshi,* Atsushi Sasaki,† Jun Abe,* Wei Qu,† Yoichiro Nakatani,† Budrul Ahsan,x Kenshiro Oshima,† Francis H. -

Lineage-Specific Evolution of the Complex Nup160 Hybrid
GENETICS | INVESTIGATION Lineage-Specific Evolution of the Complex Nup160 Hybrid Incompatibility Between Drosophila melanogaster and Its Sister Species Shanwu Tang and Daven C. Presgraves1 Department of Biology, University of Rochester, New York 14627 ABSTRACT Two genes encoding protein components of the nuclear pore complex Nup160 and Nup96 cause lethality in F2-like hybrid genotypes between Drosophila simulans and Drosophila melanogaster. In particular, D. simulans Nup160 and Nup96 each cause inviability when hemizygous or homozygous in species hybrids that are also hemizygous (or homozygous) for the D. melanogaster X chromosome. The hybrid lethality of Nup160, however, is genetically complex, depending on one or more unknown additional factors in the autosomal background. Here we study the genetics and evolution of Nup160-mediated hybrid lethality in three ways. First, we test for variability in Nup160-mediated hybrid lethality within and among the three species of the D. simulans clade— D. simulans, D. sechellia,andD. mauritiana. We show that the hybrid lethality of Nup160 is fixed in D. simulans and D. sechellia but absent in D. mauritiana. Second, we explore how the hybrid lethality of Nup160 depends on other loci in the autosomal background. We find that D. simulans Nup160-mediated hybrid lethality does not depend on the presence of D. melanogaster Nup96,andwefind that D. simulans and D. mauritiana are functionally differentiated at Nup160 as well as at other autosomal factor(s). Finally, we use population genetics data to show that Nup160 has experienced histories of recurrent positive selection both before and after the split of the three D. simulans clade species 240,000 years ago. -

Supplemental Figures
Supplemental Figures Development of a new macrophage-specific TRAP mouse (MacTRAP) and definition of the renal macrophage translational signature Andreas Hofmeister, Maximilian C. Thomassen, Sabrina Markert, André Marquardt, Mathieu Preußner, Martin Rußwurm, Ralph Schermuly, Ulrich Steinhoff, Hermann-Josef Gröne, Joachim Hoyer, Benjamin D. Humphreys, Ivica Grgic Correspondence: Ivica Grgic MD, Klinikum der Philips-Universität Marburg, Baldingerstrasse 1, 35043 Marburg. Phone: +4964215861736, email: [email protected] Sup. Fig. S1 Δ6.7fmsGFP-L10a plasmid (pGL2 backbone) Sup. Fig. S1: Plasmid map of the engineered c-fms-eGFP-L10a expression vector. Mlu1/Sal1 digestion was used for linearization and extraction of the transgene. Sup. Fig. S2 A Neutrophils Monocytes Lymphocytes B Monocytes Monocytes Neutrophils Lymphocytes H - TRAP Ly6g Count FSC CD115 Mac GFP GFP GFP GFP H - Ly6g Count FSC CD115 Wild Wild type GFP GFP GFP GFP Sup. Fig. S2: FACS analysis detects eGFP-L10a signals in monocytes but not in neutrophils or lymphocytes isolated from peripheral blood of MacTRAP mice. (A) Gating strategy to define monocyte, neutrophil and lymphocyte populations. Only single cells contributed to the analysis. (B) GFP-fluorescence was specifically detected in CD115+ monocytes, but not in Ly6g+ neutrophils or lymphocytes of MacTRAP mice. Blood samples from wild-type mice served as negative controls. Representative plots are shown, n=6. Sup. Fig. S3 A eGFP-L10a Ly6g merge+DAPI kidney 7d UUO B eGFP-L10a Ly6g merge+DAPI incision skin Tail Sup. Fig. S3: Immunostaining for mature neutrophils in fibrotic kidney tissue and tail skin biopsies from MacTRAP mice. (A) Only a very small fraction of Ly6g+ neutrophils is positive for eGFP-L10a in fibrotic kidneys after 7d UUO (0.42% ± 0.29%; 468 cells counted, n=3). -

Supplemental Figure S1 Differentially Methylated Regions (Dmrs
Supplemental Figure S1 '$$#0#,2'**7+#2&7*2#"0#%'-,11 #25##,"'1#1#122#1 '!2-0'*"#.'!2'-,-$122,1'2'-,$0-+2- !"Q !"2-$%," $ 31',% 25-$-*" !&,%# ," ' 0RTRW 1 !32V-$$ !0'2#0'T - #.0#1#,22'-, -$ "'$$#0#,2'**7+#2&7*2#"%#,#11',.0#,2#1,"2&#'0 #&4'-022,1'2'-, #25##,"'$$#0#,2"'1#1#122#1T-*!)00-51',"'!2#&7.#0+#2&7*2#"%#,#1Q%0700-51 &7.-+#2&7*2#"%#,#1Q31',%25-$-*"!&,%#,"'0RTRW1!32V-$$!0'2#0'T-%#,#1 +#22&# -4#!0'2#0'22,1'2'-,$0-+$%2-$Q5#2�#$-0#*1-',!*3"#" %#,#15'2&V4*3#0RTRWT$$#!2#"%#,10#&'%&*'%&2#" 712#0'1)1#T Supplemental Figure S2 Validation of results from the HELP assay using Epityper MassarrayT #13*21 $0-+ 2&# 1$ 117 5#0# !-00#*2#" 5'2& /3,2'22'4# +#2&7*2'-, ,*78#" 7 '13*$'2#11007$-04V-,"6U-%#,#.0-+-2#00#%'-,1T11007 51.#0$-0+#"31',%**4'* *#1+.*#1T S Supplemental Fig. S1 A unique hypermethylated genes (methylation sites) 454 (481) 5693 (6747) 120 (122) NLMGUS NEWMM REL 2963 (3207) 1338 (1560) 5 (5) unique hypomethylated genes (methylation sites) B NEWMM 0 (0) MGUS 454 (481) 0 (0) NEWMM REL NL 3* (2) 2472 (3066) NEWMM 2963 REL (3207) 2* (2) MGUS 0 (0) REL 2 (2) NEWMM 0 (0) REL Supplemental Fig. S2 A B ARID4B DNMT3A Methylation by MassArray Methylation by MassArray 0 0.2 0.4 0.6 0.8 1 1.2 0.5 0.6 0.7 0.8 0.9 1 2 0 NL PC MGUS 1.5 -0.5 NEW MM 1 REL MM -1 0.5 -1.5 0 -2 -0.5 -1 -2.5 -1.5 -3 Methylation by HELP Assay Methylation by HELP Methylation by HELP Assay Methylation by HELP -2 -3.5 -2.5 -4 Supplemental tables "3..*#+#,2*6 *#"SS 9*','!*!&0!2#0'12'!1-$.2'#,21+.*#1 DZ_STAGE Age Gender Ethnicity MM isotype PCLI Cytogenetics -

Common Differentially Expressed Genes and Pathways Correlating Both Coronary Artery Disease and Atrial Fibrillation
EXCLI Journal 2021;20:126-141– ISSN 1611-2156 Received: December 08, 2020, accepted: January 11, 2021, published: January 18, 2021 Supplementary material to: Original article: COMMON DIFFERENTIALLY EXPRESSED GENES AND PATHWAYS CORRELATING BOTH CORONARY ARTERY DISEASE AND ATRIAL FIBRILLATION Youjing Zheng, Jia-Qiang He* Department of Biomedical Sciences and Pathobiology, College of Veterinary Medicine, Virginia Tech, Blacksburg, VA 24061, USA * Corresponding author: Jia-Qiang He, Department of Biomedical Sciences and Pathobiology, Virginia Tech, Phase II, Room 252B, Blacksburg, VA 24061, USA. Tel: 1-540-231-2032. E-mail: [email protected] https://orcid.org/0000-0002-4825-7046 Youjing Zheng https://orcid.org/0000-0002-0640-5960 Jia-Qiang He http://dx.doi.org/10.17179/excli2020-3262 This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0/). Supplemental Table 1: Abbreviations used in the paper Abbreviation Full name ABCA5 ATP binding cassette subfamily A member 5 ABCB6 ATP binding cassette subfamily B member 6 (Langereis blood group) ABCB9 ATP binding cassette subfamily B member 9 ABCC10 ATP binding cassette subfamily C member 10 ABCC13 ATP binding cassette subfamily C member 13 (pseudogene) ABCC5 ATP binding cassette subfamily C member 5 ABCD3 ATP binding cassette subfamily D member 3 ABCE1 ATP binding cassette subfamily E member 1 ABCG1 ATP binding cassette subfamily G member 1 ABCG4 ATP binding cassette subfamily G member 4 ABHD18 Abhydrolase domain -

The Megalocytivirus RBIV Induces Apoptosis and MHC Class I Presentation in Rock Bream (Oplegnathus Fasciatus) Red Blood Cells
ORIGINAL RESEARCH published: 04 March 2019 doi: 10.3389/fimmu.2019.00160 The Megalocytivirus RBIV Induces Apoptosis and MHC Class I Presentation in Rock Bream (Oplegnathus fasciatus) Red Blood Cells Myung-Hwa Jung 1, Verónica Chico 2, Sergio Ciordia 3, Maria Carmen Mena 3, Sung-Ju Jung 1 and Maria Del Mar Ortega-Villaizan 2* 1 Department of Aqualife Medicine, Chonnam National University, Gwangju, South Korea, 2 IBMC-IDiBE, Universidad Miguel Hernandez, Elche, Spain, 3 Unidad de Proteómica, Centro Nacional de Biotecnología (CSIC), Madrid, Spain Rock bream iridovirus (RBIV) causes severe mass mortality in Korean rock bream (Oplegnathus fasciatus) populations. To date, immune defense mechanisms of rock bream against RBIV are unclear. While red blood cells (RBCs) are known to be involved in the immune response against viral infections, the participation of rock bream RBCs in the immune response against RBIV has not been studied yet. In this study, we examined induction of the immune response in rock bream RBCs Edited by: Brian Dixon, after RBIV infection. Each fish was injected with RBIV, and virus copy number in University of Waterloo, Canada RBCs gradually increased from 4 days post-infection (dpi), peaking at 10 dpi. A total Reviewed by: of 318 proteins were significantly regulated in RBCs from RBIV-infected individuals, Stephanie DeWitte-Orr, 183 proteins were upregulated and 135 proteins were downregulated. Differentially Wilfrid Laurier University, Canada Magdalena Chadzinska, upregulated proteins included those involved in cellular amino acid metabolic processes, Jagiellonian University, Poland cellular detoxification, snRNP assembly, and the spliceosome. Remarkably, the MHC *Correspondence: class I-related protein pathway was upregulated during RBIV infection. -
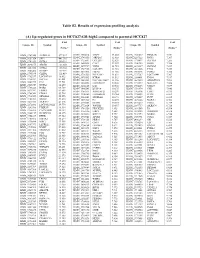
(A) Up-Regulated Genes in HCC827-GR-High2 Compared to Parental HCC827
Table S2. Results of expression profiling analysis (A) Up-regulated genes in HCC827-GR-high2 compared to parental HCC827 Fold Fold Fold Unique ID Symbol Unique ID Symbol Unique ID Symbol change* change* change* ILMN_1709348 ALDH1A1 577.587 ILMN_2310814 MAPT 13.003 ILMN_1741017 PIP4K2B 7.331 ILMN_1651354 SPP1 441.316 ILMN_1748650 MRPL45 12.988 ILMN_3237623 RNY1 7.297 ILMN_1701831 GSTA1 260.591 ILMN_1755897 UGT2B7 12.629 ILMN_1734897 SLC4A4 7.285 ILMN_1658835 CAV2 12.357 ILMN_1746359 RERG 7.280 ILMN_2094875 ABCB1 183.050 ILMN_1678939 VNN2 11.935 ILMN_1671337 SLC2A5 7.257 ILMN_3251540 GSTA2 145.982 ILMN_1729905 GAL3ST1 11.910 ILMN_1691606 LYG2 7.254 ILMN_2062468 IGFBP7 127.721 ILMN_1672536 FBLN1 11.716 ILMN_1785646 PMP22 7.246 ILMN_1795190 CLDN2 111.439 ILMN_1796339 PLEKHA2 11.631 ILMN_1737387 LOC728441 7.207 ILMN_1782937 LOC647169 98.612 ILMN_1676563 HTRA1 11.592 ILMN_1684401 FMO1 7.117 ILMN_1754247 SLC3A1 81.001 ILMN_3263423 LOC100129027 11.346 ILMN_1687035 ADAMTSL4 7.098 ILMN_1662795 CA2 79.581 ILMN_1694898 LOC653857 10.906 ILMN_2153572 MAGEA3 7.086 ILMN_2168747 GSTA2 66.250 ILMN_2404625 LAT 10.560 ILMN_1784283 USH1C 7.079 ILMN_1764228 DAB2 64.709 ILMN_1666546 DUSP14 10.375 ILMN_1731374 CPE 7.046 ILMN_1675797 EPDR1 63.605 ILMN_1764571 ARHGAP23 10.299 ILMN_1765446 EMP3 6.933 ILMN_1708341 PDZK1 59.714 ILMN_3200140 LOC645638 10.284 ILMN_1754002 IL1F8 6.863 ILMN_1713529 SEMA6A 52.575 ILMN_3244343 SNORA21 10.171 ILMN_1878007 FUT9 6.835 ILMN_1708391 NR1H4 43.218 ILMN_1671489 PC 10.075 ILMN_1699208 NAP1L1 6.763 ILMN_2412336 AKR1C2 42.826 ILMN_2404688