Genomics Reveals Alga-Associated Cyanobacteria Hiding in Plain Sight COMMENTARY John M
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

182-188 Enhanced Chlorophyll a and Primary Production in the Northern
Author version: Mar. Biol. Res., vol.8; 2012; 182-188 Enhanced chlorophyll a and primary production in the northern Arabian Sea during the spring intermonsoon due to green Noctiluca (N. scintillans) bloom N. V. Madhua,*, R. Jyothibabua, P. A. Maheswaranb, K. A. Jayaraja, C.T. Achuthankuttya aNational Institute of Oceanography, Regional Centre, Kochi -18, India bNaval Physical Oceanographic Laboratory, Kochi - 21, India Abstract The surface waters of the northeastern Arabian Sea sustained relatively high chlorophyll a (av. 0.81 ± 0.80 mgm-3) and primary production (av. 29.5 ± 23.6 mgC m-3d-1) during the early spring intermonsoon 2000. This was caused primarily by a thick patch of algal bloom spread over a vast area between 17° to 21°N and 66 to 70°E. Satellite images showed exceptionally high concentration of chlorophyll a in the bloom areas, representing the annually occurring ‘spring blooms’ during February-March. The causative organism of the bloom was the dinoflagellate, Noctiluca scintillans Macartney (synonym Noctiluca miliaris Suriray, Dinophyceae: Noctilucidea), symbiotically associated with an autotrophic prasinophyte Pedinomonas noctilucae. The symbiosis between N. scintillans and P. noctilucae is likely responsible for their explosive growth (av. 3 million cells L-1) over an extensive area making the northeastern Arabian Sea highly productive (av. 607 ± 338 mg Cm-3d-1) even during an oligotrophic period such as spring intermonsoon. Key words: - Chlorophyll a; Algal bloom; Noctiluca scintillans, Pedinomonas noctilucae; Spring intermonsoon *Email of the corresponding author - [email protected] 2 Introduction The Arabian Sea (AS hereafter) is one of the most productive regions in the Indian Ocean (Madhupratap et al., 1996), exhibiting a bimodal temperature cycles annually, with lows during winter (northeast monsoon - NEM) and summer (southwest monsoon - SWM) seasons. -

Check List 15 (5): 951–963
15 5 ANNOTATED LIST OF SPECIES Check List 15 (5): 951–963 https://doi.org/10.15560/15.5.951 Dinoflagellates in tropical estuarine waters from the Maraú River, Camamu Bay, northeastern Brazil Caio Ceza da Silva Nunes1, Sylvia Maria Moreira Susini-Ribeiro1, 2, Kaoli Pereira Cavalcante3 1 Mestrado em Sistemas Aquáticos Tropicais, Universidade Estadual de Santa Cruz, Rodovia Jorge Amado, km 16, Salobrinho, 45662090 Ilhéus, BA, Brazil. 2 Universidade Estadual de Santa Cruz, Rodovia Jorge Amado, km 16, Salobrinho, 45662090 Ilhéus, BA, Brazil. 3 Universidade Estadual Vale do Acaraú, Avenida da Universidade, 850, Campus da Betânia, Betânia, 62040370, Sobral, CE, Brazil. Corresponding author: Caio Ceza da Silva Nunes, [email protected] Abstract Dinoflagellates display great diversity in tropical regions and play an important role in the complex microbial food webs of marine and brackish environments. The goal of this study is to identify planktonic dinoflagellates and their distribution in the estuary of the Maraú River, Camamu Bay, state of Bahia, in a region with increasing use of shellfish farming. Samples were carried out monthly from August 2006 to July 2007 at four stations along the estuary. Plankton was sampled with a 20 μm mesh net. We identified 20 dinoflagellate species. The greatest species richness was ob- served in the genera Protoperidinium (five spp.), Tripos (four spp.), and Prorocentrum (three spp.). Based on literature, six species were classified as potentially harmful: Akashiwo sanguinea, Dinophysis caudata, Gonyaulax spinifera, Prorocentrum micans, Scrippsiella cf. acuminata, and Tripos furca. Protoperidinium venustum was recorded for the first time in coastal waters of Bahia. Keywords Brackish water, Dinophyta, distribution, potentially harmful species, taxonomy. -

De Rijk, L?, Caers, A,, Van De Peer, Y. & De Wachter, R. 1998. Database
BLANCHARD & HICKS-THE APICOMPLEXAN PLASTID 375 De Rijk, l?, Caers, A,, Van de Peer, Y. & De Wachter, R. 1998. Database gorad, L. & Vasil, I. K. (ed.), Cell Culture and Somatic Cell Genetics on the structure of large ribosomal subunit RNA. Nucl. Acids. Rex, of Plants, Vol7A: The molecular biology of plastids. Academic Press, 26: 183- 186. San Diego. p. 5-53. Deveraux, J., Haeberli, l? & Smithies, 0. 1984. A comprehensive set of Palmer, J. D. & Delwiche, C. E 1996. Second-hand chloroplasts and sequence analysis programs for the VAX. Nucl. Acids. Rex, 12:387-395. the case of the disappearing nucleus. Proc. Natl. Acad. Sci. USA, 93: Eaga, N. & Lang-Unnasch, N. 1995. Phylogeny of the large extrachro- 7432-7435. mosomal DNA of organisms in the phylum Apicomplexa. J. Euk. Popadic, A,, Rusch, D., Peterson, M., Rogers, B. T. & Kaufman, T. C. Microbiol,, 42:679-684. 1996. Origin of the arthropod mandible. Nature, 380:395. Fichera, M. E. & Roos, D. S. 1997. A plastid organelle as a drug target Preiser, l?, Williamson, D. H. & Wilson, R. J. M. 1995. Transfer-RNA in apicomplexan parasites. Nature, 390:407-409. genes transcribed from the plastid-like DNA of Plasmodium falci- Gardner, M. J., Williamson, D. H. & Wilson, R. J. M. 1991. A circular parum. Nucl. Acids Res., 23:4329-4336. DNA in malaria parasites encodes an RNA polymerase like that of Reith. M. & Munholland, J. 1993. A high-resolution gene map of the prokaryotes and chloroplasts. Mol. Biochem. Parasitiol., 44: 1 15-123. chloroplast genome of the red alga Porphyra purpurea. Plant Cell, Gardner, M. -

Mixotrophic Protists Among Marine Ciliates and Dinoflagellates: Distribution, Physiology and Ecology
FACULTY OF SCIENCE UNIVERSITY OF COPENHAGEN PhD thesis Woraporn Tarangkoon Mixotrophic Protists among Marine Ciliates and Dinoflagellates: Distribution, Physiology and Ecology Academic advisor: Associate Professor Per Juel Hansen Submitted: 29/04/10 Contents List of publications 3 Preface 4 Summary 6 Sammenfating (Danish summary) 8 สรุป (Thai summary) 10 The sections and objectives of the thesis 12 Introduction 14 1) Mixotrophy among marine planktonic protists 14 1.1) The role of light, food concentration and nutrients for 17 the growth of marine mixotrophic planktonic protists 1.2) Importance of marine mixotrophic protists in the 20 planktonic food web 2) Marine symbiont-bearing dinoflagellates 24 2.1) Occurrence of symbionts in the order Dinophysiales 24 2.2) The spatial distribution of symbiont-bearing dinoflagellates in 27 marine waters 2.3) The role of symbionts and phagotrophy in dinoflagellates with symbionts 28 3) Symbiosis and mixotrophy in the marine ciliate genus Mesodinium 30 3.1) Occurrence of symbiosis in Mesodinium spp. 30 3.2) The distribution of marine Mesodinium spp. 30 3.3) The role of symbionts and phagotrophy in marine Mesodinium rubrum 33 and Mesodinium pulex Conclusion and future perspectives 36 References 38 Paper I Paper II Paper III Appendix-Paper IV Appendix-I Lists of publications The thesis consists of the following papers, referred to in the synthesis by their roman numerals. Co-author statements are attached to the thesis (Appendix-I). Paper I Tarangkoon W, Hansen G Hansen PJ (2010) Spatial distribution of symbiont-bearing dinoflagellates in the Indian Ocean in relation to oceanographic regimes. Aquat Microb Ecol 58:197-213. -

Bacterial and Archaeal Symbioses with Protists, Current Biology (2021), J.Cub.2021.05.049
Please cite this article in press as: Husnik et al., Bacterial and archaeal symbioses with protists, Current Biology (2021), https://doi.org/10.1016/ j.cub.2021.05.049 ll Review Bacterial and archaeal symbioses with protists Filip Husnik1,2,*, Daria Tashyreva3, Vittorio Boscaro2, Emma E. George2, Julius Lukes3,4, and Patrick J. Keeling2,* 1Okinawa Institute of Science and Technology, Okinawa, 904-0495, Japan 2Department of Botany, University of British Columbia, Vancouver, V6T 1Z4, Canada 3Institute of Parasitology, Biology Centre, Czech Academy of Sciences, Ceske Budejovice, 370 05, Czech Republic 4Faculty of Science, University of South Bohemia, Ceske Budejovice, 370 05, Czech Republic *Correspondence: fi[email protected] (F.H.), [email protected] (P.J.K.) https://doi.org/10.1016/j.cub.2021.05.049 SUMMARY Most of the genetic, cellular, and biochemical diversity of life rests within single-celled organisms—the pro- karyotes (bacteria and archaea) and microbial eukaryotes (protists). Very close interactions, or symbioses, between protists and prokaryotes are ubiquitous, ecologically significant, and date back at least two billion years ago to the origin of mitochondria. However, most of our knowledge about the evolution and functions of eukaryotic symbioses comes from the study of animal hosts, which represent only a small subset of eukary- otic diversity. Here, we take a broad view of bacterial and archaeal symbioses with protist hosts, focusing on their evolution, ecology, and cell biology, and also explore what functions (if any) the symbionts provide to their hosts. With the immense diversity of protist symbioses starting to come into focus, we can now begin to see how these systems will impact symbiosis theory more broadly. -

The Windblown: Possible Explanations for Dinophyte DNA
bioRxiv preprint doi: https://doi.org/10.1101/2020.08.07.242388; this version posted August 10, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. The windblown: possible explanations for dinophyte DNA in forest soils Marc Gottschlinga, Lucas Czechb,c, Frédéric Mahéd,e, Sina Adlf, Micah Dunthorng,h,* a Department Biologie, Systematische Botanik und Mykologie, GeoBio-Center, Ludwig- Maximilians-Universität München, D-80638 Munich, Germany b Computational Molecular Evolution Group, Heidelberg Institute for Theoretical Studies, D- 69118 Heidelberg, Germany c Department of Plant Biology, Carnegie Institution for Science, Stanford, CA 94305, USA d CIRAD, UMR BGPI, F-34398, Montpellier, France e BGPI, Université de Montpellier, CIRAD, IRD, Montpellier SupAgro, Montpellier, France f Department of Soil Sciences, College of Agriculture and Bioresources, University of Saskatchewan, Saskatoon, S7N 5A8, SK, Canada g Eukaryotic Microbiology, Faculty of Biology, Universität Duisburg-Essen, D-45141 Essen, Germany h Centre for Water and Environmental Research (ZWU), Universität Duisburg-Essen, D- 45141 Essen, Germany Running title: Dinophytes in soils Correspondence M. Dunthorn, Eukaryotic Microbiology, Faculty of Biology, Universität Duisburg-Essen, Universitätsstrasse 5, D-45141 Essen, Germany Telephone number: +49-(0)-201-183-2453; email: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/2020.08.07.242388; this version posted August 10, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -
Diversity.Pdf
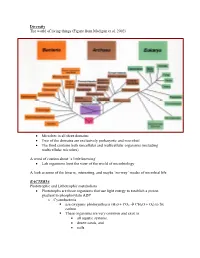
Diversity The world of living things (Figure from Madigan et al. 2002) • Microbes in all three domains • Two of the domains are exclusively prokaryotic and microbial • The third contains both unicellular and multicellular organisms (including multicellular microbes) A word of caution about ‘a little knowing’ • Lab organisms limit the view of the world of microbiology A look at some of the bizarre, interesting, and maybe ‘no-way’ modes of microbial life BACTERIA Phototrophic and Lithotrophic metabolism • Phototrophs are those organisms that use light energy to establish a proton gradient to phosphorylate ADP o Cyanobacteria use oxygenic photosynthesis (H20 + CO2 Æ CH2O + O2) to fix carbon. These organisms are very common and exist in • all aquatic systems, • desert sands, and • soils. Many have the ability to fix elemental N2 into NH3 and this only enhances their ability to survive in diverse habitats Why is this important? (Table-Figures from Madigan et al. 2002) Figure 14.38 o Phototrophic bacteria Use anoxygenic photosynthesis (H2S + CO2 Æ CH2O + S2) to fix carbon into cell stuff Primarily found in restricted habitats • H2S must be available • Light must be available • Located in ‘bands in lakes and near the surface of muddy environments Some may grow chemoorganotrophically in the dark Some may grow photoheterotrophically (Figures from Madigan et al. 2002) • Chemolithotrophs o Organisms that obtain energy from oxidation of inorganic compounds o Most can fix CO2 into cell stuff using the Calvin Cycle (that is the one green plants use) o Many can obtain reduced coenzymes by operating electron transport chains in reverse. Nitrifying bacteria • Grow at the expense of reduced inorganic nitrogen and all oxidations are aerobic. -

Molecular Phylogeny of Selected Species of the Order Dinophysiales (Dinophyceae)—Testing the Hypothesis of a Dinophysioid Radiation1
J. Phycol. 45, 1136–1152 (2009) Ó 2009 Phycological Society of America DOI: 10.1111/j.1529-8817.2009.00741.x MOLECULAR PHYLOGENY OF SELECTED SPECIES OF THE ORDER DINOPHYSIALES (DINOPHYCEAE)—TESTING THE HYPOTHESIS OF A DINOPHYSIOID RADIATION1 Maria Hastrup Jensen and Niels Daugbjerg2 Phycology Laboratory, Department of Biology, University of Copenhagen, Øster Farimagsgade 2D, DK-1353 Copenhagen K, Denmark Almost 80 years ago, a radiation scheme based on Abbreviations: bp, base pairs; LSL, left sulcal lists; structural resemblance was first outlined for the ML, maximum likelihood; pp, posterior probabil- marine order Dinophysiales. This hypothetical radia- ities; R1–3, ribs 1–3 tion illustrated the relationship between the dino- physioid genera and included several independent, extant lineages. Subsequent studies have supplied Members of the dinoflagellate order Dinophysi- additional information on morphology and ecology ales are distributed worldwide in the marine envi- to these evolutionary lineages. We have for the first ronment. However, a vast majority of the nearly time combined morphological information with 300 recognized species are found in tropical waters molecular phylogenies to test the dinophysioid radi- (Kofoid and Skogsberg 1928, Taylor 1976, Gomez ation hypothesis in a modern context. Nuclear- 2005a). Even though the dinophysioids seldom are encoded LSU rDNA sequences including domains abundant in numbers, incidents of severe seasonal D1-D6 from 27 species belonging to Dinophysis blooms of Dinophysis species have been recorded in Ehrenb., Ornithocercus F. Stein, Phalacroma F. Stein, some areas (Kofoid and Skogsberg 1928, Maestrine Amphisolenia F. Stein, Citharistes F. Stein, and et al. 1996, Guillou et al. 2002, Gomez 2007). Pro- Histioneis F. -

Scrippsiella Trochoidea (F.Stein) A.R.Loebl
MOLECULAR DIVERSITY AND PHYLOGENY OF THE CALCAREOUS DINOPHYTES (THORACOSPHAERACEAE, PERIDINIALES) Dissertation zur Erlangung des Doktorgrades der Naturwissenschaften (Dr. rer. nat.) der Fakultät für Biologie der Ludwig-Maximilians-Universität München zur Begutachtung vorgelegt von Sylvia Söhner München, im Februar 2013 Erster Gutachter: PD Dr. Marc Gottschling Zweiter Gutachter: Prof. Dr. Susanne Renner Tag der mündlichen Prüfung: 06. Juni 2013 “IF THERE IS LIFE ON MARS, IT MAY BE DISAPPOINTINGLY ORDINARY COMPARED TO SOME BIZARRE EARTHLINGS.” Geoff McFadden 1999, NATURE 1 !"#$%&'(&)'*!%*!+! +"!,-"!'-.&/%)$"-"!0'* 111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111 2& ")3*'4$%/5%6%*!+1111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111 7! 8,#$0)"!0'*+&9&6"*,+)-08!+ 111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111 :! 5%*%-"$&0*!-'/,)!0'* 11111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111 ;! "#$!%"&'(!)*+&,!-!"#$!'./+,#(0$1$!2! './+,#(0$1$!-!3+*,#+4+).014!1/'!3+4$0&41*!041%%.5.01".+/! 67! './+,#(0$1$!-!/&"*.".+/!1/'!4.5$%"(4$! 68! ./!5+0&%!-!"#$!"#+*10+%,#1$*10$1$! 69! "#+*10+%,#1$*10$1$!-!5+%%.4!1/'!$:"1/"!'.;$*%."(! 6<! 3+4$0&41*!,#(4+)$/(!-!0#144$/)$!1/'!0#1/0$! 6=! 1.3%!+5!"#$!"#$%.%! 62! /0+),++0'* 1111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111111<=! -

<I>Ornithocercus Magnificus</I>
THECAL MORPHOLOGY OF ORNITHOCERCUS MAGNIFI- CUS (D1NOFLAGELLATA) WITH NOTES ON RELATED SPECIESl DEAN R. NORRIS Department of Oceanography, Texas A&M University, College Station, Texas 77843 ABSTRACT Detailed descriptions of the thecal plates of Omithocercus magnificus are given. Though a few differences in structural detail are disclosed, the number and general arrangement of these plates are the same as those of Omithocercus thumi. Species in the family Ornithocercidae which are rare or previously unreported from the Gulf of Mexico include six in the genus Omithocercus, eight in Histioneis and one in Parahistioneis. Figures are included for all species found. Synonymies and general descriptions are given for those which differ from previously described specimens. INTRODUCTION The identification of many dinoflagellate species is somewhat problem- atic. Armored forms of dinoflagellates have complex thecae which provide several morphological characters valuable in taxonomic studies. The most fundamental taxonomic features are the number and arrangement of the thecal plates. Shape of the body, lists (membranes), spines, and similar characters are of secondary importance (Graham, 1942: 6). Relationships within the group are based on thecal structures, yet relatively few papers are available with a critical analysis of the thecal plates. Little was known of the thecal morphology in the tribe Dinophysoidae until the study of Tai & Skogsberg (1934). They established that it is considerably more complex than was previously assumed. Their work was limited to the family Dinophysidae. According to Tai & Skogsberg (1934: 387-88), several earlier workers (SchUtt, Kofoid, & Calkins) indicated fewer plates than are actually present. Although Schlitt illustrated plates in the sulcal region he did not discuss them. -

Downloaded from the EMBL-EBI Repository Biostudies
bioRxiv preprint doi: https://doi.org/10.1101/2021.05.28.446125; this version posted May 29, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. A robust approach to estimate relative phytoplankton cell abundance from metagenomic data Juan José Pierella Karlusich1,2 *, Eric Pelletier2,3, Lucie Zinger1, Fabien Lombard2,4,5, Adriana Zingone6, Sébastien Colin7,8,9, Josep M. Gasol10, Richard Dorrell1, Eleonora Scalco6, Silvia G. Acinas10, Patrick Wincker2,3, Colomban de Vargas2,8, Chris Bowler1,2 * 1 Institut de Biologie de l'ENS (IBENS), Département de biologie, École normale supérieure, CNRS, INSERM, Université PSL, 75005 Paris, France 2 CNRS Research Federation for the study of Global Ocean Systems Ecology and Evolution, FR2022/Tara Oceans GOSEE, 3 rue Michel-Ange, 75016 Paris, France 3 Génomique Métabolique, Genoscope, Institut François Jacob, CEA, CNRS, Univ Evry, Université Paris-Saclay, 91057 Evry, France 4 Sorbonne Universités, CNRS, Laboratoire d'Océanographie de Villefranche (LOV), 06230 Villefranche-sur-Mer, France 5 Institut Universitaire de France (IUF), Paris, France 6 Stazione Zoologica Anton Dohrn, Villa Comunale, 80121 Naples, Italy 7 European Molecular Biology Laboratory, Heidelberg, Germany 8 Sorbonne Université, CNRS, Station Biologique de Roscoff, UMR 7144, ECOMAP, 29680 Roscoff, France 9 Max Planck Institute for Developmental Biology, Tübingen, Germany 10 Department of Marine Biology and Oceanography, Institut de Ciènces del Mar, CSIC, Barcelona, Spain *corresponding authors: Juan José Pierella Karlusich ([email protected]) and Chris Bowler ([email protected]) Running title: phytoplankton relative abundance from metagenomes Keywords: photosynthesis, phytoplankton, psbO, V9-18S rRNA, metabarcoding, metagenomics, metatranscriptomics, Tara Oceans bioRxiv preprint doi: https://doi.org/10.1101/2021.05.28.446125; this version posted May 29, 2021. -

Physico-Chemical and Biological Factors Influencing Dinoflagellate
Biogeosciences, 15, 2325–2348, 2018 https://doi.org/10.5194/bg-15-2325-2018 © Author(s) 2018. This work is distributed under the Creative Commons Attribution 4.0 License. Physico-chemical and biological factors influencing dinoflagellate cyst production in the Cariaco Basin Manuel Bringué1,2,a, Robert C. Thunell1, Vera Pospelova2, James L. Pinckney1,3, Oscar E. Romero4, and Eric J. Tappa1 1School of the Earth, Ocean and Environment, University of South Carolina, 701 Sumter Street, EWS 617, Columbia, SC 29208, USA 2School of Earth and Ocean Sciences, University of Victoria, P.O. Box 1700, STN CSC, Victoria, BC, V8W 2Y2, Canada 3Belle W. Baruch Institute for Marine and Coastal Sciences, University of South Carolina, 700 Sumter Street, EWS 604, Columbia, SC 29208, USA 4MARUM, Center for Marine Environmental Sciences, University Bremen, Leobenerstraße, 28359 Bremen, Germany anow at: Geological Survey of Canada, 3303 33rd Street NW, Calgary, AB, T2L 2A7, Canada Correspondence: Manuel Bringué ([email protected]) Received: 20 November 2017 – Discussion started: 22 November 2017 Revised: 19 March 2018 – Accepted: 25 March 2018 – Published: 19 April 2018 Abstract. We present a 2.5-year-long sediment trap record duction in the basin with a 1-year lag, and may have con- of dinoflagellate cyst production in the Cariaco Basin, off tributed to the unusually high fluxes of cysts type “Cp” Venezuela (southern Caribbean Sea). The site lies under (possibly the cysts of the toxic dinoflagellate Cochlodinium the influence of wind-driven, seasonal upwelling which pro- polykrikoides sensu Li et al., 2015), with cyst type Cp fluxes motes high levels of primary productivity during boreal win- up to 11.8 × 103 cysts m−2 day−1 observed during the weak ter and spring.