De Rijk, L?, Caers, A,, Van De Peer, Y. & De Wachter, R. 1998. Database
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

182-188 Enhanced Chlorophyll a and Primary Production in the Northern
Author version: Mar. Biol. Res., vol.8; 2012; 182-188 Enhanced chlorophyll a and primary production in the northern Arabian Sea during the spring intermonsoon due to green Noctiluca (N. scintillans) bloom N. V. Madhua,*, R. Jyothibabua, P. A. Maheswaranb, K. A. Jayaraja, C.T. Achuthankuttya aNational Institute of Oceanography, Regional Centre, Kochi -18, India bNaval Physical Oceanographic Laboratory, Kochi - 21, India Abstract The surface waters of the northeastern Arabian Sea sustained relatively high chlorophyll a (av. 0.81 ± 0.80 mgm-3) and primary production (av. 29.5 ± 23.6 mgC m-3d-1) during the early spring intermonsoon 2000. This was caused primarily by a thick patch of algal bloom spread over a vast area between 17° to 21°N and 66 to 70°E. Satellite images showed exceptionally high concentration of chlorophyll a in the bloom areas, representing the annually occurring ‘spring blooms’ during February-March. The causative organism of the bloom was the dinoflagellate, Noctiluca scintillans Macartney (synonym Noctiluca miliaris Suriray, Dinophyceae: Noctilucidea), symbiotically associated with an autotrophic prasinophyte Pedinomonas noctilucae. The symbiosis between N. scintillans and P. noctilucae is likely responsible for their explosive growth (av. 3 million cells L-1) over an extensive area making the northeastern Arabian Sea highly productive (av. 607 ± 338 mg Cm-3d-1) even during an oligotrophic period such as spring intermonsoon. Key words: - Chlorophyll a; Algal bloom; Noctiluca scintillans, Pedinomonas noctilucae; Spring intermonsoon *Email of the corresponding author - [email protected] 2 Introduction The Arabian Sea (AS hereafter) is one of the most productive regions in the Indian Ocean (Madhupratap et al., 1996), exhibiting a bimodal temperature cycles annually, with lows during winter (northeast monsoon - NEM) and summer (southwest monsoon - SWM) seasons. -

Check List 15 (5): 951–963
15 5 ANNOTATED LIST OF SPECIES Check List 15 (5): 951–963 https://doi.org/10.15560/15.5.951 Dinoflagellates in tropical estuarine waters from the Maraú River, Camamu Bay, northeastern Brazil Caio Ceza da Silva Nunes1, Sylvia Maria Moreira Susini-Ribeiro1, 2, Kaoli Pereira Cavalcante3 1 Mestrado em Sistemas Aquáticos Tropicais, Universidade Estadual de Santa Cruz, Rodovia Jorge Amado, km 16, Salobrinho, 45662090 Ilhéus, BA, Brazil. 2 Universidade Estadual de Santa Cruz, Rodovia Jorge Amado, km 16, Salobrinho, 45662090 Ilhéus, BA, Brazil. 3 Universidade Estadual Vale do Acaraú, Avenida da Universidade, 850, Campus da Betânia, Betânia, 62040370, Sobral, CE, Brazil. Corresponding author: Caio Ceza da Silva Nunes, [email protected] Abstract Dinoflagellates display great diversity in tropical regions and play an important role in the complex microbial food webs of marine and brackish environments. The goal of this study is to identify planktonic dinoflagellates and their distribution in the estuary of the Maraú River, Camamu Bay, state of Bahia, in a region with increasing use of shellfish farming. Samples were carried out monthly from August 2006 to July 2007 at four stations along the estuary. Plankton was sampled with a 20 μm mesh net. We identified 20 dinoflagellate species. The greatest species richness was ob- served in the genera Protoperidinium (five spp.), Tripos (four spp.), and Prorocentrum (three spp.). Based on literature, six species were classified as potentially harmful: Akashiwo sanguinea, Dinophysis caudata, Gonyaulax spinifera, Prorocentrum micans, Scrippsiella cf. acuminata, and Tripos furca. Protoperidinium venustum was recorded for the first time in coastal waters of Bahia. Keywords Brackish water, Dinophyta, distribution, potentially harmful species, taxonomy. -

Genomics Reveals Alga-Associated Cyanobacteria Hiding in Plain Sight COMMENTARY John M
COMMENTARY Genomics reveals alga-associated cyanobacteria hiding in plain sight COMMENTARY John M. Archibalda,b,1 Cyanobacteria occupy a special place in the pantheon of prokaryotic life. It is in the ancestors of these ubiquitous microbes that oxygenic photosynthesis first evolved more than 2 billion y ago (1), and it is from endosymbiotic cyanobacteria that the plastids (chloro- plasts) of plants and algae are derived (2). Modern-day cyanobacteria are diverse in form and function; they in- clude coccoid marine picoplankton such as Prochloro- coccus (3), freshwater biofilm-forming genera [e.g., Gloeomargarita (4)], and filamentous taxa capable of fix- ing nitrogen [e.g., Nostoc (5)]. In PNAS, Nakayama et al. (6) add an exciting chapter to the story of cyanobacterial diversity. The authors describe the genome sequence of a cyanobacterium living ectosymbiotically on an eye- catching dinoflagellate named Ornithocercus magnifi- cus. Their results provide insight into the nature of an enigmatic symbiotic relationship and reveal the exis- tence of a cryptic, globally distributed cyanobacterial lineage that has until now gone unappreciated. Ornithocercus is indeed magnificent, even by di- Fig. 1. Light micrograph of an Ornithocercus noflagellate standards. The surface of this heterotro- dinoflagellate. The ectosymbiotic OmCyn cyanobacteria phic marine protist is decorated with crown-shaped, are visible within an extracellular chamber on the cellulosic outcroppings that extend from the cell body “upper” crown of the cell. Image courtesy of Takuro Nakayama (Tohoku University, Sendai, Japan). in different directions (Fig. 1) (7). The “upper” crown forms an extracellular chamber in which autofluores- cent cyanobacteria reside, and microscopic evidence genome of the organism, which they dubbed “OmCyn.” suggests that the bacteria can be vertically transmitted In a preliminary phylogenetic analysis of the 16S ribo- from mother to daughter chambers during host cell somal RNA (rRNA) gene, OmCyn was found to branch division (8). -

Protocols for Monitoring Harmful Algal Blooms for Sustainable Aquaculture and Coastal Fisheries in Chile (Supplement Data)
Protocols for monitoring Harmful Algal Blooms for sustainable aquaculture and coastal fisheries in Chile (Supplement data) Provided by Kyoko Yarimizu, et al. Table S1. Phytoplankton Naming Dictionary: This dictionary was constructed from the species observed in Chilean coast water in the past combined with the IOC list. Each name was verified with the list provided by IFOP and online dictionaries, AlgaeBase (https://www.algaebase.org/) and WoRMS (http://www.marinespecies.org/). The list is subjected to be updated. Phylum Class Order Family Genus Species Ochrophyta Bacillariophyceae Achnanthales Achnanthaceae Achnanthes Achnanthes longipes Bacillariophyta Coscinodiscophyceae Coscinodiscales Heliopeltaceae Actinoptychus Actinoptychus spp. Dinoflagellata Dinophyceae Gymnodiniales Gymnodiniaceae Akashiwo Akashiwo sanguinea Dinoflagellata Dinophyceae Gymnodiniales Gymnodiniaceae Amphidinium Amphidinium spp. Ochrophyta Bacillariophyceae Naviculales Amphipleuraceae Amphiprora Amphiprora spp. Bacillariophyta Bacillariophyceae Thalassiophysales Catenulaceae Amphora Amphora spp. Cyanobacteria Cyanophyceae Nostocales Aphanizomenonaceae Anabaenopsis Anabaenopsis milleri Cyanobacteria Cyanophyceae Oscillatoriales Coleofasciculaceae Anagnostidinema Anagnostidinema amphibium Anagnostidinema Cyanobacteria Cyanophyceae Oscillatoriales Coleofasciculaceae Anagnostidinema lemmermannii Cyanobacteria Cyanophyceae Oscillatoriales Microcoleaceae Annamia Annamia toxica Cyanobacteria Cyanophyceae Nostocales Aphanizomenonaceae Aphanizomenon Aphanizomenon flos-aquae -

Mixotrophic Protists Among Marine Ciliates and Dinoflagellates: Distribution, Physiology and Ecology
FACULTY OF SCIENCE UNIVERSITY OF COPENHAGEN PhD thesis Woraporn Tarangkoon Mixotrophic Protists among Marine Ciliates and Dinoflagellates: Distribution, Physiology and Ecology Academic advisor: Associate Professor Per Juel Hansen Submitted: 29/04/10 Contents List of publications 3 Preface 4 Summary 6 Sammenfating (Danish summary) 8 สรุป (Thai summary) 10 The sections and objectives of the thesis 12 Introduction 14 1) Mixotrophy among marine planktonic protists 14 1.1) The role of light, food concentration and nutrients for 17 the growth of marine mixotrophic planktonic protists 1.2) Importance of marine mixotrophic protists in the 20 planktonic food web 2) Marine symbiont-bearing dinoflagellates 24 2.1) Occurrence of symbionts in the order Dinophysiales 24 2.2) The spatial distribution of symbiont-bearing dinoflagellates in 27 marine waters 2.3) The role of symbionts and phagotrophy in dinoflagellates with symbionts 28 3) Symbiosis and mixotrophy in the marine ciliate genus Mesodinium 30 3.1) Occurrence of symbiosis in Mesodinium spp. 30 3.2) The distribution of marine Mesodinium spp. 30 3.3) The role of symbionts and phagotrophy in marine Mesodinium rubrum 33 and Mesodinium pulex Conclusion and future perspectives 36 References 38 Paper I Paper II Paper III Appendix-Paper IV Appendix-I Lists of publications The thesis consists of the following papers, referred to in the synthesis by their roman numerals. Co-author statements are attached to the thesis (Appendix-I). Paper I Tarangkoon W, Hansen G Hansen PJ (2010) Spatial distribution of symbiont-bearing dinoflagellates in the Indian Ocean in relation to oceanographic regimes. Aquat Microb Ecol 58:197-213. -

Molecular Phylogeny of Sinophysis: Evaluating the Possible Early 2 3 Q1 Evolutionary History of Dinophysoid Dinoflagellates 4 5 M
1 Molecular phylogeny of Sinophysis: Evaluating the possible early 2 3 Q1 evolutionary history of dinophysoid dinoflagellates 4 5 M. HOPPENRATH1,2*, N. CHOME´ RAT3 & B. LEANDER2 6 1 7 Forschungsinstitut Senckenberg, Deutsches Zentrum fu¨r Marine Biodiversita¨tsforschung 8 (DZMB), Su¨dstrand 44, D-26382 Wilhelmshaven, Germany 9 2 10 Departments of Zoology and Botany, University of British Columbia, Canadian Institute 11 for Advanced Research, Program in Integrated Microbial Biodiversity, Vancouver, 12 BC, V67 1Z4, Canada 13 3 14 IFREMER, LER FBN Station de Concarneau, 13 rue de Ke´rose, 29187 15 Concarneau Cedex, France 16 17 *Corresponding author (e-mail: [email protected]) 18 19 20 Abstract: Dinophysoids are a group of thecate dinoflagellates with a very distinctive thecal plate 21 arrangement involving a sagittal suture: the so-called dinophysoid tabulation pattern. Although the number and layout of the thecal plates is highly conserved, the morphological diversity within the 22 group is outstandingly high for dinoflagellates. Previous hypotheses about character evolution 23 within dinophysoids based on comparative morphology alone are currently being evaluated by 24 molecular phylogenetic studies. Sinophysis is especially significant within the context of these 25 hypotheses because several features within this genus approximate the inferred ancestral states 26 for dinophysoids as a whole, such as a (benthic) sand-dwelling lifestyle, a relatively streamlined 27 theca and a heterotrophic mode of nutrition. We generated and analysed small subunit (SSU) 28 rDNA sequences for five different species of Sinophysis, including the type species (S. ebriola, 29 S. stenosoma, S. grandis, S. verruculosa and S. microcephala). We also generated SSU rDNA 30 sequences from the planktonic dinophysoid Oxyphysis (O. -

Phytoplankton Biomass and Composition in a Well-Flushed, Sub-Tropical Estuary: the Contrasting Effects of Hydrology, Nutrient Lo
Marine Environmental Research 112 (2015) 9e20 Contents lists available at ScienceDirect Marine Environmental Research journal homepage: www.elsevier.com/locate/marenvrev Phytoplankton biomass and composition in a well-flushed, sub-tropical estuary: The contrasting effects of hydrology, nutrient loads and allochthonous influences * J.A. Hart a, E.J. Phlips a, , S. Badylak a, N. Dix b, K. Petrinec b, A.L. Mathews c, W. Green d, A. Srifa a a Fisheries and Aquatic Sciences Program, SFRC, University of Florida, 7922 NW 71st Street, Gainesville, FL 32653, USA b Guana Tolomato Matanzas National Estuarine Research Reserve, 505 Guana River Road, Ponte Vedra, FL 32082, USA c Georgia Southern University, Department of Biology, Statesboro, GA 30460, USA d St. Johns River Water Management District, 4049 Reid Street, Palatka, FL 32177, USA article info abstract Article history: The primary objective of this study was to examine trends in phytoplankton biomass and species Received 10 March 2015 composition under varying nutrient load and hydrologic regimes in the Guana Tolomato Matanzas es- Received in revised form tuary (GTM), a well-flushed sub-tropical estuary located on the northeast coast of Florida. The GTM 28 July 2015 contains both regions of significant human influence and pristine areas with only modest development, Accepted 31 August 2015 providing a test case for comparing and contrasting phytoplankton community dynamics under varying Available online 4 September 2015 degrees of nutrient load. Water temperature, salinity, Secchi disk depth, nutrient concentrations and chlorophyll concentrations were determined on a monthly basis from 2002 to 2012 at three represen- Keywords: Guana Tolomato Matanzas estuary tative sampling sites in the GTM. -

Bacterial and Archaeal Symbioses with Protists, Current Biology (2021), J.Cub.2021.05.049
Please cite this article in press as: Husnik et al., Bacterial and archaeal symbioses with protists, Current Biology (2021), https://doi.org/10.1016/ j.cub.2021.05.049 ll Review Bacterial and archaeal symbioses with protists Filip Husnik1,2,*, Daria Tashyreva3, Vittorio Boscaro2, Emma E. George2, Julius Lukes3,4, and Patrick J. Keeling2,* 1Okinawa Institute of Science and Technology, Okinawa, 904-0495, Japan 2Department of Botany, University of British Columbia, Vancouver, V6T 1Z4, Canada 3Institute of Parasitology, Biology Centre, Czech Academy of Sciences, Ceske Budejovice, 370 05, Czech Republic 4Faculty of Science, University of South Bohemia, Ceske Budejovice, 370 05, Czech Republic *Correspondence: fi[email protected] (F.H.), [email protected] (P.J.K.) https://doi.org/10.1016/j.cub.2021.05.049 SUMMARY Most of the genetic, cellular, and biochemical diversity of life rests within single-celled organisms—the pro- karyotes (bacteria and archaea) and microbial eukaryotes (protists). Very close interactions, or symbioses, between protists and prokaryotes are ubiquitous, ecologically significant, and date back at least two billion years ago to the origin of mitochondria. However, most of our knowledge about the evolution and functions of eukaryotic symbioses comes from the study of animal hosts, which represent only a small subset of eukary- otic diversity. Here, we take a broad view of bacterial and archaeal symbioses with protist hosts, focusing on their evolution, ecology, and cell biology, and also explore what functions (if any) the symbionts provide to their hosts. With the immense diversity of protist symbioses starting to come into focus, we can now begin to see how these systems will impact symbiosis theory more broadly. -

The Windblown: Possible Explanations for Dinophyte DNA
bioRxiv preprint doi: https://doi.org/10.1101/2020.08.07.242388; this version posted August 10, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. The windblown: possible explanations for dinophyte DNA in forest soils Marc Gottschlinga, Lucas Czechb,c, Frédéric Mahéd,e, Sina Adlf, Micah Dunthorng,h,* a Department Biologie, Systematische Botanik und Mykologie, GeoBio-Center, Ludwig- Maximilians-Universität München, D-80638 Munich, Germany b Computational Molecular Evolution Group, Heidelberg Institute for Theoretical Studies, D- 69118 Heidelberg, Germany c Department of Plant Biology, Carnegie Institution for Science, Stanford, CA 94305, USA d CIRAD, UMR BGPI, F-34398, Montpellier, France e BGPI, Université de Montpellier, CIRAD, IRD, Montpellier SupAgro, Montpellier, France f Department of Soil Sciences, College of Agriculture and Bioresources, University of Saskatchewan, Saskatoon, S7N 5A8, SK, Canada g Eukaryotic Microbiology, Faculty of Biology, Universität Duisburg-Essen, D-45141 Essen, Germany h Centre for Water and Environmental Research (ZWU), Universität Duisburg-Essen, D- 45141 Essen, Germany Running title: Dinophytes in soils Correspondence M. Dunthorn, Eukaryotic Microbiology, Faculty of Biology, Universität Duisburg-Essen, Universitätsstrasse 5, D-45141 Essen, Germany Telephone number: +49-(0)-201-183-2453; email: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/2020.08.07.242388; this version posted August 10, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -
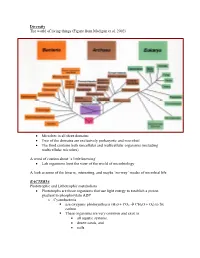
Diversity.Pdf
Diversity The world of living things (Figure from Madigan et al. 2002) • Microbes in all three domains • Two of the domains are exclusively prokaryotic and microbial • The third contains both unicellular and multicellular organisms (including multicellular microbes) A word of caution about ‘a little knowing’ • Lab organisms limit the view of the world of microbiology A look at some of the bizarre, interesting, and maybe ‘no-way’ modes of microbial life BACTERIA Phototrophic and Lithotrophic metabolism • Phototrophs are those organisms that use light energy to establish a proton gradient to phosphorylate ADP o Cyanobacteria use oxygenic photosynthesis (H20 + CO2 Æ CH2O + O2) to fix carbon. These organisms are very common and exist in • all aquatic systems, • desert sands, and • soils. Many have the ability to fix elemental N2 into NH3 and this only enhances their ability to survive in diverse habitats Why is this important? (Table-Figures from Madigan et al. 2002) Figure 14.38 o Phototrophic bacteria Use anoxygenic photosynthesis (H2S + CO2 Æ CH2O + S2) to fix carbon into cell stuff Primarily found in restricted habitats • H2S must be available • Light must be available • Located in ‘bands in lakes and near the surface of muddy environments Some may grow chemoorganotrophically in the dark Some may grow photoheterotrophically (Figures from Madigan et al. 2002) • Chemolithotrophs o Organisms that obtain energy from oxidation of inorganic compounds o Most can fix CO2 into cell stuff using the Calvin Cycle (that is the one green plants use) o Many can obtain reduced coenzymes by operating electron transport chains in reverse. Nitrifying bacteria • Grow at the expense of reduced inorganic nitrogen and all oxidations are aerobic. -

Cell Lysis of a Phagotrophic Dinoflagellate, Polykrikos Kofoidii Feeding on Alexandrium Tamarense
Plankton Biol. Ecol. 47 (2): 134-136,2000 plankton biology & ecology f The Plankton Society of Japan 2(100 Note Cell lysis of a phagotrophic dinoflagellate, Polykrikos kofoidii feeding on Alexandrium tamarense Hyun-Jin Cho1 & Kazumi Matsuoka2 'Graduate School of Marine Science and Engineering, Nagasaki University, 1-14 Bunkyo-inachi, Nagasaki 852-8521. Japan 2Laboratory of Coastal Environmental Sciences, Faculty of Fisheries. Nagasaki University. 1-14 Bunkyo-machi, Nagasaki 852-8521. Japan Received 9 December 1999; accepted 10 April 2000 In many cases, phytoplankton blooms terminate suddenly dinoflagellate, Alexandrium tamarense (Lebour) Balech. within a few days. For bloom-forming phytoplankton, grazing P. kofoidii was collected from Isahaya Bay in western is one of the major factors in the decline of blooms as is sexual Kyushu, Japan, on 3 November, 1998. We isolated actively reproduction to produce non-dividing gametes and planozy- swimming P. kofoidii using a capillary pipette and individually gotes (Anderson et al. 1983; Frost 1991). Matsuoka et al. transferred them into multi-well tissue culture plates contain (2000) reported growth rates of a phagotrophic dinoflagellate, ing a dense suspension of the autotrophic dinoflagellate, Polykrikos kofoidii Chatton, using several dinoflagellate Gymnodinium catenatum Graham (approximately 700 cells species as food organisms. They noted that P. kofoidii showed ml"1) isolated from a bloom near Amakusa Island, western various feeding and growth responses to strains of Alexan Japan, 1997. The P. kofoidii were cultured at 20°C with con drium and Prorocentrum. This fact suggests that for ecological stant lighting to a density of 60 indiv. ml"1, and then starved control of phytoplankton blooms, we should collect infor until no G. -

Molecular Phylogeny of Selected Species of the Order Dinophysiales (Dinophyceae)—Testing the Hypothesis of a Dinophysioid Radiation1
J. Phycol. 45, 1136–1152 (2009) Ó 2009 Phycological Society of America DOI: 10.1111/j.1529-8817.2009.00741.x MOLECULAR PHYLOGENY OF SELECTED SPECIES OF THE ORDER DINOPHYSIALES (DINOPHYCEAE)—TESTING THE HYPOTHESIS OF A DINOPHYSIOID RADIATION1 Maria Hastrup Jensen and Niels Daugbjerg2 Phycology Laboratory, Department of Biology, University of Copenhagen, Øster Farimagsgade 2D, DK-1353 Copenhagen K, Denmark Almost 80 years ago, a radiation scheme based on Abbreviations: bp, base pairs; LSL, left sulcal lists; structural resemblance was first outlined for the ML, maximum likelihood; pp, posterior probabil- marine order Dinophysiales. This hypothetical radia- ities; R1–3, ribs 1–3 tion illustrated the relationship between the dino- physioid genera and included several independent, extant lineages. Subsequent studies have supplied Members of the dinoflagellate order Dinophysi- additional information on morphology and ecology ales are distributed worldwide in the marine envi- to these evolutionary lineages. We have for the first ronment. However, a vast majority of the nearly time combined morphological information with 300 recognized species are found in tropical waters molecular phylogenies to test the dinophysioid radi- (Kofoid and Skogsberg 1928, Taylor 1976, Gomez ation hypothesis in a modern context. Nuclear- 2005a). Even though the dinophysioids seldom are encoded LSU rDNA sequences including domains abundant in numbers, incidents of severe seasonal D1-D6 from 27 species belonging to Dinophysis blooms of Dinophysis species have been recorded in Ehrenb., Ornithocercus F. Stein, Phalacroma F. Stein, some areas (Kofoid and Skogsberg 1928, Maestrine Amphisolenia F. Stein, Citharistes F. Stein, and et al. 1996, Guillou et al. 2002, Gomez 2007). Pro- Histioneis F.