Xenopus ADAM19 Regulates Wnt Signaling and Neural Crest
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplemental Information to Mammadova-Bach Et Al., “Laminin Α1 Orchestrates VEGFA Functions in the Ecosystem of Colorectal Carcinogenesis”
Supplemental information to Mammadova-Bach et al., “Laminin α1 orchestrates VEGFA functions in the ecosystem of colorectal carcinogenesis” Supplemental material and methods Cloning of the villin-LMα1 vector The plasmid pBS-villin-promoter containing the 3.5 Kb of the murine villin promoter, the first non coding exon, 5.5 kb of the first intron and 15 nucleotides of the second villin exon, was generated by S. Robine (Institut Curie, Paris, France). The EcoRI site in the multi cloning site was destroyed by fill in ligation with T4 polymerase according to the manufacturer`s instructions (New England Biolabs, Ozyme, Saint Quentin en Yvelines, France). Site directed mutagenesis (GeneEditor in vitro Site-Directed Mutagenesis system, Promega, Charbonnières-les-Bains, France) was then used to introduce a BsiWI site before the start codon of the villin coding sequence using the 5’ phosphorylated primer: 5’CCTTCTCCTCTAGGCTCGCGTACGATGACGTCGGACTTGCGG3’. A double strand annealed oligonucleotide, 5’GGCCGGACGCGTGAATTCGTCGACGC3’ and 5’GGCCGCGTCGACGAATTCACGC GTCC3’ containing restriction site for MluI, EcoRI and SalI were inserted in the NotI site (present in the multi cloning site), generating the plasmid pBS-villin-promoter-MES. The SV40 polyA region of the pEGFP plasmid (Clontech, Ozyme, Saint Quentin Yvelines, France) was amplified by PCR using primers 5’GGCGCCTCTAGATCATAATCAGCCATA3’ and 5’GGCGCCCTTAAGATACATTGATGAGTT3’ before subcloning into the pGEMTeasy vector (Promega, Charbonnières-les-Bains, France). After EcoRI digestion, the SV40 polyA fragment was purified with the NucleoSpin Extract II kit (Machery-Nagel, Hoerdt, France) and then subcloned into the EcoRI site of the plasmid pBS-villin-promoter-MES. Site directed mutagenesis was used to introduce a BsiWI site (5’ phosphorylated AGCGCAGGGAGCGGCGGCCGTACGATGCGCGGCAGCGGCACG3’) before the initiation codon and a MluI site (5’ phosphorylated 1 CCCGGGCCTGAGCCCTAAACGCGTGCCAGCCTCTGCCCTTGG3’) after the stop codon in the full length cDNA coding for the mouse LMα1 in the pCIS vector (kindly provided by P. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Conservation and Divergence of ADAM Family Proteins in the Xenopus Genome
Wei et al. BMC Evolutionary Biology 2010, 10:211 http://www.biomedcentral.com/1471-2148/10/211 RESEARCH ARTICLE Open Access ConservationResearch article and divergence of ADAM family proteins in the Xenopus genome Shuo Wei*1, Charles A Whittaker2, Guofeng Xu1, Lance C Bridges1,3, Anoop Shah1, Judith M White1 and Douglas W DeSimone1 Abstract Background: Members of the disintegrin metalloproteinase (ADAM) family play important roles in cellular and developmental processes through their functions as proteases and/or binding partners for other proteins. The amphibian Xenopus has long been used as a model for early vertebrate development, but genome-wide analyses for large gene families were not possible until the recent completion of the X. tropicalis genome sequence and the availability of large scale expression sequence tag (EST) databases. In this study we carried out a systematic analysis of the X. tropicalis genome and uncovered several interesting features of ADAM genes in this species. Results: Based on the X. tropicalis genome sequence and EST databases, we identified Xenopus orthologues of mammalian ADAMs and obtained full-length cDNA clones for these genes. The deduced protein sequences, synteny and exon-intron boundaries are conserved between most human and X. tropicalis orthologues. The alternative splicing patterns of certain Xenopus ADAM genes, such as adams 22 and 28, are similar to those of their mammalian orthologues. However, we were unable to identify an orthologue for ADAM7 or 8. The Xenopus orthologue of ADAM15, an active metalloproteinase in mammals, does not contain the conserved zinc-binding motif and is hence considered proteolytically inactive. We also found evidence for gain of ADAM genes in Xenopus as compared to other species. -

View / Download 3.3 Mb
Identification of Mechanisms and Pathways Involved in MLL2-Mediated Tumorigenesis by Chun-Chi Chang Department of Pathology Duke University Date:_______________________ Approved: ___________________________ Yiping He, Supervisor ___________________________ Salvatore Pizzo ___________________________ Hai Yan Thesis submitted in partial fulfillment of the requirements for the degree of Master of Science in the Department of Pathology in the Graduate School of Duke University 2013 ABSTRACT Identification of Mechanisms and Pathways Involved in MLL2-Mediated Tumorigenesis by Chun-Chi Chang Department of Pathology Duke University Date:_______________________ Approved: ___________________________ Yiping He, Supervisor ___________________________ Salvatore Pizzo ___________________________ Hai Yan An abstract of a thesis submitted in partial fulfillment of the requirements for the degree of Master of Science in the Department of Pathology in the Graduate School of Duke University 2013 Copyright by Chun-Chi Chang 2013 Abstract Myeloid/lymphoid or mixed-lineage leukemia (MLL)-family genes encode histone lysine methyltransferases that play important roles in epigenetic regulation of gene transcription, and these genes are frequently mutated in human cancers. While MLL1 and MLL4 have been the most extensively studied, MLL2 and its homolog MLL3 are not well-understood. Specifically, little is known regarding the extent of global MLL2 involvement in the regulation of gene expression and the mechanism underlying its alterations in mediating tumorigenesis. To study the role of MLL2 in tumorigenesis, we somatically knocked out MLL2 in a colorectal carcinoma cell line, HCT116. We observed that the MLL2 loss of function results in significant reduction of cell growth and multinuclear morphology. We further profiled MLL2 regulated genes and pathways by analyzing gene expression in MLL2 wild-type versus MLL2-null isogenic cell lines. -

Fibroblasts from the Human Skin Dermo-Hypodermal Junction Are
cells Article Fibroblasts from the Human Skin Dermo-Hypodermal Junction are Distinct from Dermal Papillary and Reticular Fibroblasts and from Mesenchymal Stem Cells and Exhibit a Specific Molecular Profile Related to Extracellular Matrix Organization and Modeling Valérie Haydont 1,*, Véronique Neiveyans 1, Philippe Perez 1, Élodie Busson 2, 2 1, 3,4,5,6, , Jean-Jacques Lataillade , Daniel Asselineau y and Nicolas O. Fortunel y * 1 Advanced Research, L’Oréal Research and Innovation, 93600 Aulnay-sous-Bois, France; [email protected] (V.N.); [email protected] (P.P.); [email protected] (D.A.) 2 Department of Medical and Surgical Assistance to the Armed Forces, French Forces Biomedical Research Institute (IRBA), 91223 CEDEX Brétigny sur Orge, France; [email protected] (É.B.); [email protected] (J.-J.L.) 3 Laboratoire de Génomique et Radiobiologie de la Kératinopoïèse, Institut de Biologie François Jacob, CEA/DRF/IRCM, 91000 Evry, France 4 INSERM U967, 92260 Fontenay-aux-Roses, France 5 Université Paris-Diderot, 75013 Paris 7, France 6 Université Paris-Saclay, 78140 Paris 11, France * Correspondence: [email protected] (V.H.); [email protected] (N.O.F.); Tel.: +33-1-48-68-96-00 (V.H.); +33-1-60-87-34-92 or +33-1-60-87-34-98 (N.O.F.) These authors contributed equally to the work. y Received: 15 December 2019; Accepted: 24 January 2020; Published: 5 February 2020 Abstract: Human skin dermis contains fibroblast subpopulations in which characterization is crucial due to their roles in extracellular matrix (ECM) biology. -
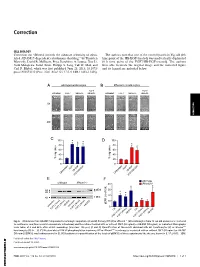
Irhom2 Controls the Substrate Selectivity of Stimulated ADAM17-Dependent Ectodomain Shedding
Correction CELL BIOLOGY Correction for “iRhom2 controls the substrate selectivity of stimu- The authors note that one of the control panels in Fig. 6B (0 h lated ADAM17-dependent ectodomain shedding,” by Thorsten time point of the HB-EGF-treated) was inadvertently duplicated Maretzky, David R. McIlwain, Priya Darshinee A. Issuree, Xue Li, (0 h time point of the FGF7/HB-EGF-treated). The authors Jordi Malapeira, Sadaf Amin, Philipp A. Lang, Tak W. Mak, and were able to locate the original image and the corrected figure Carl P. Blobel, which was first published June 25, 2013; 10.1073/ and its legend are included below. pnas.1302553110 (Proc. Natl. Acad. Sci. U.S.A. 110,11433–11438). AB CORRECTION C D E F Fig. 6. iRhom2 controls ADAM17-dependent keratinocyte migration. (A and B) Primary WT (A)oriRhom2−/− (B) keratinocytes from 12-wk-old animals were cultured to confluence, and then a scratch wound was introduced, and the cultures treated with or without FGF7 (50 ng/mL) or HB-EGF (50 ng/mL), as indicated. Micrographs − − were taken at 0 and 48 h after scratch wounding. (Scale bar: 100 μm.) (C and D) Quantification of the results obtained with WT keratinocytes (C)oriRhom2 / − − keratinocytes (D)(n = 3). (E) Western blot of ERK1/2 phosphorylation in primary WT or iRhom2 / keratinocytes incubated with or without FGF7 (20 ng/mL) or HB-EGF (50 ng/mL) (ERK1/2 was loading control in E). (F) Densitometric quantification of the levels of pERK1/2 of three experiments like the one shown in E.*P ≤ 0.05; ±SEM. -

The Adams Family of Metalloproteases: Multidomain Proteins with Multiple Functions
Downloaded from genesdev.cshlp.org on September 26, 2021 - Published by Cold Spring Harbor Laboratory Press REVIEW The ADAMs family of metalloproteases: multidomain proteins with multiple functions Darren F. Seals and Sara A. Courtneidge1 Van Andel Research Institute, Grand Rapids, Michigan 49503, USA The ADAMs family of transmembrane proteins belongs diseases such as arthritis and cancer (Chang and Werb to the zinc protease superfamily. Members of the family 2001). Adamalysins are similar to the matrixins in their have a modular design, characterized by the presence of metalloprotease domains, but contain a unique integrin metalloprotease and integrin receptor-binding activities, receptor-binding disintegrin domain (Fig. 1). It is the and a cytoplasmic domain that in many family members presence of these two domains that give the ADAMs specifies binding sites for various signal transducing pro- their name (a disintegrin and metalloprotease). The do- teins. The ADAMs family has been implicated in the main structure of the ADAMs consists of a prodomain, a control of membrane fusion, cytokine and growth factor metalloprotease domain, a disintegrin domain, a cyste- shedding, and cell migration, as well as processes such as ine-rich domain, an EGF-like domain, a transmembrane muscle development, fertilization, and cell fate determi- domain, and a cytoplasmic tail. The adamalysins sub- nation. Pathologies such as inflammation and cancer family also contains the class III snake venom metallo- also involve ADAMs family members. Excellent reviews proteases and the ADAM-TS family, which although covering various facets of the ADAMs literature-base similar to the ADAMs, can be distinguished structurally have been published over the years and we recommend (Fig. -

Hsa-Mir-100-5P, an Overexpressed Mirna in Human Ovarian
Takebayashi et al. Reproductive Biology and Endocrinology (2020) 18:31 https://doi.org/10.1186/s12958-020-00590-3 RESEARCH Open Access hsa-miR-100-5p, an overexpressed miRNA in human ovarian endometriotic stromal cells, promotes invasion through attenuation of SMARCD1 expression Kanetoshi Takebayashi1, Kaei Nasu1,2*, Mamiko Okamoto1, Yoko Aoyagi1, Tomoko Hirakawa1 and Hisashi Narahara1 Abstract Background: A number of microRNAs are aberrantly expressed in endometriosis and are involved in its pathogenesis. Our previous study demonstrated that has-miR-100-5p expression is enhanced in human endometriotic cyst stromal cells (ECSCs). The present study aimed to elucidate the roles of has-miR-100-5p in the pathogenesis of endometriosis. Methods: Normal endometrial stromal cells (NESCs) were isolated from normal eutopic endometrium without endometriosis. Using hsa-miR-100-5p-transfected NESCs, we evaluated the effect of hsa-miR-100-5p on the invasiveness of these cells by Transwell invasion assay and in-vitro wound repair assay. We also investigated the downstream signal pathways of hsa-miR-100-5p by microarray analysis and Ingenuity pathways analysis. Results: hsa-miR-100-5p transfection enhanced the invasion and motility of NESCs. After hsa-miR-100-5p transfection, mRNA expression of SWItch/sucrose non-fermentable-related matrix-associated actin-dependent regulator of chromatin subfamily D member 1 (SMARCD1) was significantly attenuated. Whereas, the expression of matrix metallopeptidase 1 (MMP1) mRNA and active MMP1 protein levels was upregulated. Conclusion: We found that SMARCD1/MMP-1 is a downstream pathway of hsa-miR-100-5p. hsa-miR-100-5p transfection enhanced the motility of NESCs by inhibiting SMARCD1 expression and MMP1 activation. -

ADAM8 As a Drug Target in Pancreatic Cancer
ARTICLE Received 19 Feb 2014 | Accepted 24 Dec 2014 | Published 28 Jan 2015 DOI: 10.1038/ncomms7175 ADAM8 as a drug target in pancreatic cancer Uwe Schlomann1,2, Garrit Koller1, Catharina Conrad2, Taheera Ferdous1, Panagiota Golfi1, Adolfo Molejon Garcia1, Sabrina Ho¨fling1, Maddy Parsons3, Patricia Costa4, Robin Soper4, Maud Bossard4, Thorsten Hagemann4, Rozita Roshani4, Norbert Sewald5, Randal R. Ketchem6, Marcia L. Moss7, Fred H. Rasmussen7, Miles A. Miller8, Douglas A. Lauffenburger8, David A. Tuveson9, Christopher Nimsky2 &Jo¨rg W. Bartsch1,2 Pancreatic ductal adenocarcinoma (PDAC) has a grim prognosis with o5% survivors after 5 years. High expression levels of ADAM8, a metalloprotease disintegrin, are correlated with poor clinical outcome. We show that ADAM8 expression is associated with increased migration and invasiveness of PDAC cells caused by activation of ERK1/2 and higher MMP activities. For biological function, ADAM8 requires multimerization and associates with b1 integrin on the cell surface. A peptidomimetic ADAM8 inhibitor, BK-1361, designed by structural modelling of the disintegrin domain, prevents ADAM8 multimerization. In PDAC cells, BK-1361 affects ADAM8 function leading to reduced invasiveness, and less ERK1/2 and MMP activation. BK-1361 application in mice decreased tumour burden and metastasis of implanted pancreatic tumour cells and provides improved metrics of clinical symptoms and survival in a KrasG12D-driven mouse model of PDAC. Thus, our data integrate ADAM8 in pancreatic cancer signalling and validate ADAM8 as a target for PDAC therapy. 1 King’s College London, Institute for Pharmaceutical Science and KCLDI, London SE1 9RT, UK. 2 Department of Neurosurgery, Marburg University, , Baldingerstrasse, 35033 Marburg, Germany. -

Autocrine IFN Signaling Inducing Profibrotic Fibroblast Responses By
Downloaded from http://www.jimmunol.org/ by guest on September 23, 2021 Inducing is online at: average * The Journal of Immunology , 11 of which you can access for free at: 2013; 191:2956-2966; Prepublished online 16 from submission to initial decision 4 weeks from acceptance to publication August 2013; doi: 10.4049/jimmunol.1300376 http://www.jimmunol.org/content/191/6/2956 A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Autocrine IFN Signaling Feng Fang, Kohtaro Ooka, Xiaoyong Sun, Ruchi Shah, Swati Bhattacharyya, Jun Wei and John Varga J Immunol cites 49 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html http://www.jimmunol.org/content/suppl/2013/08/20/jimmunol.130037 6.DC1 This article http://www.jimmunol.org/content/191/6/2956.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2013 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 23, 2021. The Journal of Immunology A Synthetic TLR3 Ligand Mitigates Profibrotic Fibroblast Responses by Inducing Autocrine IFN Signaling Feng Fang,* Kohtaro Ooka,* Xiaoyong Sun,† Ruchi Shah,* Swati Bhattacharyya,* Jun Wei,* and John Varga* Activation of TLR3 by exogenous microbial ligands or endogenous injury-associated ligands leads to production of type I IFN. -

ADAM19: a Novel Target for Metabolic Syndrome in Humans and Mice
University of Texas Rio Grande Valley ScholarWorks @ UTRGV School of Medicine Publications and Presentations School of Medicine 2017 ADAM19: A Novel Target for Metabolic Syndrome in Humans and Mice Lakshini Weerasekera Caroline Rudnicka Qing-Xiang Sang Joanne E. Curran The University of Texas Rio Grande Valley Matthew P. Johnson The University of Texas Rio Grande Valley See next page for additional authors Follow this and additional works at: https://scholarworks.utrgv.edu/som_pub Part of the Medicine and Health Sciences Commons Recommended Citation Weerasekera, L., Rudnicka, C., Sang, Q.-X., Curran, J. E., Johnson, M. P., Moses, E. K., Göring, H. H. H., Blangero, J., Hricova, J., Schlaich, M., & Matthews, V. B. (2017). ADAM19: A Novel Target for Metabolic Syndrome in Humans and Mice. Mediators of Inflammation, 2017, 7281986. https://doi.org/10.1155/ 2017/7281986 This Article is brought to you for free and open access by the School of Medicine at ScholarWorks @ UTRGV. It has been accepted for inclusion in School of Medicine Publications and Presentations by an authorized administrator of ScholarWorks @ UTRGV. For more information, please contact [email protected], [email protected]. Authors Lakshini Weerasekera, Caroline Rudnicka, Qing-Xiang Sang, Joanne E. Curran, Matthew P. Johnson, Eric K. Moses, Harald H. H. Goring, John Blangero, Jana Hricova, Markus Schlaich, and Vance B. Matthews This article is available at ScholarWorks @ UTRGV: https://scholarworks.utrgv.edu/som_pub/123 Hindawi Mediators of Inflammation Volume 2017, Article ID 7281986, 9 pages https://doi.org/10.1155/2017/7281986 Research Article ADAM19: A Novel Target for Metabolic Syndrome in Humans and Mice Lakshini Weerasekera,1 Caroline Rudnicka,2 Qing-Xiang Sang,3 Joanne E. -

University of Groningen the Biology of Adams in Renal Disease
University of Groningen The biology of ADAMs in renal disease Melenhorst, Wynand Bernhard Willem Henderik IMPORTANT NOTE: You are advised to consult the publisher's version (publisher's PDF) if you wish to cite from it. Please check the document version below. Document Version Publisher's PDF, also known as Version of record Publication date: 2009 Link to publication in University of Groningen/UMCG research database Citation for published version (APA): Melenhorst, W. B. W. H. (2009). The biology of ADAMs in renal disease. s.n. Copyright Other than for strictly personal use, it is not permitted to download or to forward/distribute the text or part of it without the consent of the author(s) and/or copyright holder(s), unless the work is under an open content license (like Creative Commons). Take-down policy If you believe that this document breaches copyright please contact us providing details, and we will remove access to the work immediately and investigate your claim. Downloaded from the University of Groningen/UMCG research database (Pure): http://www.rug.nl/research/portal. For technical reasons the number of authors shown on this cover page is limited to 10 maximum. Download date: 27-09-2021 2 ADAMALYSINS IN BIOLOGY AND DISEASE Harry van Goor1 Wynand Melenhorst1 Anthony Turner2 Stephen Holgate3 1. Department of Pathology and Medical Biology, University Medical Center Groningen, University of Groningen, the Netherlands 2. Division of Infection, Inflammation and Repair, Southampton General Hospital, Southampton, United Kingdom 3. Institute of Molecular and Cellular Biology, Faculty of Biological Sciences, University of Leeds, Leeds, United Kingdom.