Structural and Functional Analysis of the Vaccinia Virus O1 Virulence Protein
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Poxviruses: Smallpox Vaccine, Its Complications and Chemotherapy
Virus Adaptation and Treatment Dovepress open access to scientific and medical research Open Access Full Text Article R E V IEW Poxviruses: smallpox vaccine, its complications and chemotherapy Mimi Remichkova Abstract: The threat of bioterrorism in the recent years has once again posed to mankind the unresolved problems of contagious diseases, well forgotten in the past. Smallpox (variola) is Department of Pathogenic Bacteria, The Stephan Angeloff Institute among the most dangerous and highly contagious viral infections affecting humans. The last of Microbiology, Bulgarian Academy natural case in Somalia marked the end of a successful World Health Organization campaign of Sciences, Sofia, Bulgaria for smallpox eradication by vaccination on worldwide scale. Smallpox virus still exists today in some laboratories, specially designated for that purpose. The contemporary response in the treatment of the post-vaccine complications, which would occur upon enforcing new programs for mass-scale smallpox immunization, includes application of effective chemotherapeutics and their combinations. The goals are to provide the highest possible level of protection and safety of For personal use only. the population in case of eventual terrorist attack. This review describes the characteristic features of the poxviruses, smallpox vaccination, its adverse reactions, and poxvirus chemotherapy. Keywords: poxvirus, smallpox vaccine, post vaccine complications, inhibitors Characteristics of poxviruses Smallpox (variola) infection is caused by the smallpox virus. This virus belongs to the genus of Orthopoxvirus included in the Poxviridae family. Poxviruses are one of the largest and most complexly structured viruses, known so far. The genome of poxviruses consists of a linear two-chained DNA and its replication takes place in the cytoplasm of the infected cell. -
Generalized Vaccinia After Smallpox Vaccination with Concomitant Primary Epstein Barr Virus Infection
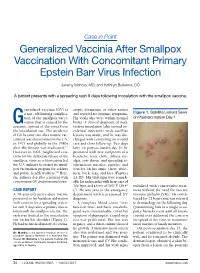
Case in Point Generalized Vaccinia After Smallpox Vaccination With Concomitant Primary Epstein Barr Virus Infection Jeremy Mandia, MD; and Kathryn Buikema, DO A patient presents with a spreading rash 9 days following inoculation with the smallpox vaccine. eneralized vaccinia (GV) is atopic dermatitis, or other rashes a rare, self-limiting complica- and reported no systemic symptoms. Figure 1. Satellite Lesions Seen tion of the smallpox vacci- His vitals also were within normal on Postvaccination Day 1 Gnation that is caused by the limits. A clinical diagnosis of inad- systemic spread of the virus from vertent inoculation (also termed ac- the inoculation site. The incidence cidental infection) with satellite of GV became rare after routine vac- lesions was made, and he was dis- cination was discontinued in the U.S. charged with counseling on wound in 1971 and globally in the 1980s care and close follow-up. Two days after the disease was eradicated.1,2 later, on postvaccination day 11, he However in 2002, heightened con- presented with new symptoms of a cerns for the deliberate release of the headache, fever, chills, diffuse my- smallpox virus as a bioweapon led algia, sore throat, and spreading er- the U.S. military to restart its small- ythematous macules, papules, and pox vaccination program for soldiers vesicles on his arms, chest, abdo- and public health workers.3,4 Here, men, back, legs, and face (Figures the authors describe a patient with 2A-2D). His vital signs were remark- concomitant GV and mononucleosis. able for tachycardia with heart rate of 100 bpm and a fever of 103º F (39.4º stabilized with conservative treat- CASE REPORT C). -

Medical Management of Biological Casualties Handbook
USAMRIID’s MEDICAL MANAGEMENT OF BIOLOGICAL CASUALTIES HANDBOOK Sixth Edition April 2005 U.S. ARMY MEDICAL RESEARCH INSTITUTE OF INFECTIOUS DISEASES FORT DETRICK FREDERICK, MARYLAND Emergency Response Numbers National Response Center: 1-800-424-8802 or (for chem/bio hazards & terrorist events) 1-202-267-2675 National Domestic Preparedness Office: 1-202-324-9025 (for civilian use) Domestic Preparedness Chem/Bio Helpline: 1-410-436-4484 or (Edgewood Ops Center – for military use) DSN 584-4484 USAMRIID’s Emergency Response Line: 1-888-872-7443 CDC'S Emergency Response Line: 1-770-488-7100 Handbook Download Site An Adobe Acrobat Reader (pdf file) version of this handbook can be downloaded from the internet at the following url: http://www.usamriid.army.mil USAMRIID’s MEDICAL MANAGEMENT OF BIOLOGICAL CASUALTIES HANDBOOK Sixth Edition April 2005 Lead Editor Lt Col Jon B. Woods, MC, USAF Contributing Editors CAPT Robert G. Darling, MC, USN LTC Zygmunt F. Dembek, MS, USAR Lt Col Bridget K. Carr, MSC, USAF COL Ted J. Cieslak, MC, USA LCDR James V. Lawler, MC, USN MAJ Anthony C. Littrell, MC, USA LTC Mark G. Kortepeter, MC, USA LTC Nelson W. Rebert, MS, USA LTC Scott A. Stanek, MC, USA COL James W. Martin, MC, USA Comments and suggestions are appreciated and should be addressed to: Operational Medicine Department Attn: MCMR-UIM-O U.S. Army Medical Research Institute of Infectious Diseases (USAMRIID) Fort Detrick, Maryland 21702-5011 PREFACE TO THE SIXTH EDITION The Medical Management of Biological Casualties Handbook, which has become affectionately known as the "Blue Book," has been enormously successful - far beyond our expectations. -

Vaccinia Virus
APPENDIX 2 Vaccinia Virus • Accidental infection following transfer from the vac- cination site to another site (autoinoculation) or to Disease Agent: another person following intimate contact Likelihood of Secondary Transmission: • Vaccinia virus • Significant following direct contact Disease Agent Characteristics: At-Risk Populations: • Family: Poxviridae; Subfamily: Chordopoxvirinae; • Individuals receiving smallpox (vaccinia) vaccination Genus: Orthopoxvirus • Individuals who come in direct contact with vacci- • Virion morphology and size: Enveloped, biconcave nated persons core with two lateral bodies, brick-shaped to pleo- • Those at risk for more severe complications of infec- morphic virions, ~360 ¥ 270 ¥ 250 nm in size tion include the following: • Nucleic acid: Nonsegmented, linear, covalently ᭺ Immune-compromised persons including preg- closed, double-stranded DNA, 18.9-20.0 kb in length nant women • Physicochemical properties: Virus is inactivated at ᭺ Patients with atopy, especially those with eczema 60°C for 8 minutes, but antigen can withstand 100°C; ᭺ Patients with extensive exfoliative skin disease lyophilized virus maintains potency for 18 months at 4-6°C; virus may be stable when dried onto inanimate Vector and Reservoir Involved: surfaces; susceptible to 1% sodium hypochlorite, • No natural host 2% glutaraldehyde, and formaldehyde; disinfection of hands and environmental contamination with soap Blood Phase: and water are effective • Vaccinia DNA was detected by PCR in the blood in 6.5% of 77 military members from 1 to 3 weeks after Disease Name: smallpox (vaccinia) vaccination that resulted in a major skin reaction. • Progressive vaccinia (vaccinia necrosum or vaccinia • In the absence of complications after immunization, gangrenosum) recently published PCR and culture data suggest that • Generalized vaccinia viremia with current vaccines must be rare 3 weeks • Eczema vaccinatum after vaccination. -

(12) Patent Application Publication (10) Pub. No.: US 2009/0221523 A1 Tseng Et Al
US 2009.022 1523A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2009/0221523 A1 Tseng et al. (43) Pub. Date: Sep. 3, 2009 (54) NORTH-2'-DEOXY-METHANO- (86). PCT No.: PCT/US2O06/02O894 CARBATHYMIDNES AS ANTIVIRAL AGENTS AGAINST POXVIRUSES S371 (c)(1), (2), (4) Date: Apr. 3, 2009 (76) Inventors: Christopher K. Tseng, Related U.S. Application Data Burtonsville, MD (US); Victor E. (60) Provisional application No. 60/684.811, filed on May Marquez, Montgomery Village, 25, 2005. MD (US) Publication Classification (51) Int. Cl. Correspondence Address: A 6LX 3L/7072 (2006.01) KNOBBE, MARTENS, OLSON & BEAR, LLP A6IP3L/20 (2006.01) 2040 MAINSTREET, FOURTEENTH FLOOR (52) U.S. Cl. .......................................................... S14/SO IRVINE, CA 92.614 (US) (57) ABSTRACT (21) Appl. No.: 11/920,881 A method for the prevention or treatment of poxvirus infec tion by administering an effective amount of an antiviral agent comprising cyclopropanated carbocyclic 2'-deoxy (22) PCT Filed: May 25, 2006 nucleoside to an individual in need thereof is provided. Patent Application Publication Sep. 3, 2009 Sheet 2 of 3 US 2009/022 1523 A1 HSV-2 TK Goalpox TK humonl TK VV TK VOriod TK monkeypox TK fowlpox TK HSV-1 K african swine fever virus TK VZV TK EBV TK FIF2 Patent Application Publication Sep. 3, 2009 Sheet 3 of 3 US 2009/022 1523 A1 CC (N)-MCT CDV VC – -2 48.61 site - 2322 2027 . US 2009/022 1523 A1 Sep. 3, 2009 NORTH-2-DEOXY. breaks depended on the isolation of infected individuals and METHANOCARBATHYMIDNES AS the vaccination of close contacts. -

(Vaccinia) Adverse Reactions
Morbidity and Mortality Weekly Report Recommendations and Reports February 3, 2006 / Vol. 55 / No. RR-1 Surveillance Guidelines for Smallpox Vaccine (vaccinia) Adverse Reactions INSIDE: Continuing Education Examination department of health and human services Centers for Disease Control and Prevention MMWR CONTENTS The MMWR series of publications is published by the Introduction......................................................................... 1 Coordinating Center for Health Information and Service, Centers for Disease Control and Prevention (CDC), U.S. Reporting Guidelines ........................................................... 2 Department of Health and Human Services, Atlanta, GA 30333. Case Definition and Classification ....................................... 2 Localized Reactions ........................................................... 3 SUGGESTED CITATION Unintentional Transfer of Vaccinia Virus ............................ 3 Centers for Disease Control and Prevention. Surveillance guidelines for smallpox vaccine (vaccinia) adverse reactions. Diffuse Dermatologic Complications ................................. 4 MMWR 2006;55(No. RR-1):[inclusive page numbers]. Progressive Vaccinia ......................................................... 6 Rare Reactions .................................................................. 6 Centers for Disease Control and Prevention Cardiac ............................................................................ 7 Julie L. Gerberding, MD, MPH Case Classification ........................................................... -

Developing Drugs to Mitigate Complications from Smallpox Vaccine
Guidance for Industry Vaccinia Virus — Developing Drugs to Mitigate Complications from Smallpox Vaccination DRAFT GUIDANCE This guidance document is being distributed for comment purposes only. Comments and suggestions regarding this draft document should be submitted within 60 days of publication in the Federal Register of the notice announcing the availability of the draft guidance. Submit comments to Division of Dockets Management (HFA-305), Food and Drug Administration, 5630 Fishers Lane, rm. 1061, Rockville, MD 20852. All comments should be identified with the docket number listed in the notice of availability that publishes in the Federal Register. For questions regarding this draft document contact (CDER) Lewis Schrager 301-827-7711 or Debra B. Birnkrant 301-827-2330, (CBER) Stephen Ripley 301-827-6210, or (CDRH) Steve Gutman 301-594-3084. U.S. Department of Health and Human Services Food and Drug Administration Center for Drug Evaluation and Research (CDER) Center for Biologics Evaluation and Research (CBER) Center for Devices and Radiological Health (CDRH) March 2004 Clinical Antimicrobial Guidance for Industry Vaccinia Virus — Developing Drugs to Mitigate Complications from Smallpox Vaccination Additional copies are available from: Office of Training and Communication Division of Drug Information, HFD-240 Center for Drug Evaluation and Research Food and Drug Administration 5600 Fishers Lane Rockville, MD 20857 (Tel) 301-827-4573 http://www.fda.gov/cder/guidance/index.htm or Office of Communication, Training and Manufacturers Assistance, HFM-40 Center for Biologics Evaluation and Research Food and Drug Administration 1401 Rockville Pike, Rockville, MD 20852-1448 http://www.fda.gov/cber/guidelines.htm Phone: the Voice Information System at 800-835-4709 or 301-827-1800 or Office of Health and Industry Programs Division of Small Manufacturers Assistance (HFZ-220) 1350 Piccard Drive Rockville, MD 20850-4307 U.S.A. -

Kaposi's Varicelliform Eruption
CONTINUING MEDICAL EDUCATION Kaposi’s Varicelliform Eruption: A Case Report and Review of the Literature Sasha C. Kramer, MD; Chadwick J. Thomas, MD; William B. Tyler, MD; Dirk M. Elston, MD GOAL To gain a thorough understanding of Kaposi’s varicelliform eruption OBJECTIVES Upon completion of this activity, dermatologists and general practitioners should be able to: 1. Recognize Kaposi’s varicelliform eruption. 2. Distinguish herpetic from vaccine-associated forms of Kaposi’s varicelliform eruption. 3. Manage Kaposi’s varicelliform eruption in affected patients. CME Test on page 123. This article has been peer reviewed and is accredited by the ACCME to provide continuing approved by Michael Fisher, MD, Professor of medical education for physicians. Medicine, Albert Einstein College of Medicine. Albert Einstein College of Medicine designates Review date: January 2004. this educational activity for a maximum of 1 This activity has been planned and implemented category 1 credit toward the AMA Physician’s in accordance with the Essential Areas and Policies Recognition Award. Each physician should claim of the Accreditation Council for Continuing Medical only that hour of credit that he/she actually spent Education through the joint sponsorship of Albert in the activity. Einstein College of Medicine and Quadrant This activity has been planned and produced in HealthCom, Inc. Albert Einstein College of Medicine accordance with ACCME Essentials. Drs. Kramer, Thomas, Tyler, and Elston report no conflict of interest. The authors report discussion of off-label use for intravenous vaccinia immune globulin under an investigational new drug protocol. Dr. Fisher reports no conflict of interest. Disseminated herpes or vaccinia in the setting of eczema herpeticum in a woman with a long- underlying skin diseases is known as Kaposi’s standing history of atopic dermatitis (AD). -

ICD-9-CM Tabular Addenda Cdc-Pdf
ICD-9-CM TABULAR ADDENDA October 1, 2008 (FY09) The official version of ICD-9-CM on CD-ROM has been modified to indicate which codes have a required fifth-digit. The applicable fifth-digits are included in brackets below the code. An addenda of these changes is in a separate PDF file, available via a link from the NCHS web page related to the ICD-9-CM addenda, below. http://www.cdc.gov/nchs/datawh/ftpserv/ftpicd9/ftpicd9.htm#addenda 006 Amebiasis 006.8 Amebic infection of other sites Revise Excludes: specific infections by free-living amebae (136.21- 136.29) Revise POLIOMYELITIS AND OTHER NON-ARTHROPOD-BORNE VIRAL DISEASES AND PRION DISEASES OF CENTRAL NERVOUS SYSTEM (045-049) 038 Septicemia 038.1 Staphylococcal septicemia Revise 038.11 Methicillin susceptible Staphylococcus aureus septicemia Add MSSA septicemia Add Staphylococcus aureus septicemia NOS New code 038.12 Methicillin resistant Staphylococcus aureus septicemia 041 Bacterial infection in conditions classified elsewhere and of unspecified site 041.1 Staphylococcus Revise 041.11 Methicillin susceptible Staphylococcus aureus Add MSSA Add Staphylococcus aureus NOS New code 041.12 Methicillin resistant Staphylococcus aureus Methicillin-resistant staphylococcus aureus (MRSA) 1 ICD-9-CM Tabular Addenda Key: Underline: added text October 1, 2008 (FY09) Strikeout: deleted text Revise 046 Slow virus infections and prion diseases of central nervous system 046.1 Jakob-Creutzfeldt disease Delete Subacute spongiform encephalopathy New code 046.11 Variant Creutzfeldt-Jakob disease vCJD New -

Boards Fodder Vaccines in Dermatology by Caroline A
boards fodder Vaccines in dermatology By Caroline A. Nelson, MD LIVE INACTIVATED/KILLED TOXOID SUBUNIT/ CONJUGATE Adenovirus Hepatitis A virus Diphtheria, tetanus, and Anthrax Cholera (oral) Influenza (injection) pertussis Haemophilus influenzae Influenza (intranasal) Japanese encephalitis Hepatitis B virus Measles, mumps, rubella Polio (injection) Human papillomavirus Polio (oral) Rabies Influenza (injection) Rotavirus Meningococcus Smallpox Pneumococcus Tuberculosis Typhoid (injection) Typhoid (oral) Zoster (Shingrix) Varicella zoster virus Yellow fever Zoster (Zostavax) VACCINE ROUTINE INDICATIONS SKIN REACTIONS/ COMPLICATIONS* NOTES† Viral Infections Hepatitis B Infants at 0-, 2-, and 6-months of Anetoderma, granuloma annulare, lichen Patients without evidence of disease or immunity virus (HBV) age and at-risk adults planus, lichen nitidus, lichen striatus, on serologic testing and with risk factors should papular acrodermatitis of childhood be offered vaccination prior to immunosuppression (Gianotti-Crosti syndrome), polyarteritis nodosa, and pseudolymphoma Human papil- Gardasil and Gardasil-9: Patients Localized lipoatrophy Vaccines contain L1 capsid protein of specific HPV lomavirus 9-26 years of age with a second types: Cervarix has 16 and 18; Gardasil has 6, 11, (HPV) dose after 6-12 months (patients 16, and 18; and Gardasil-9 has 6, 11, 16, 18, 31, 33, 15-26 years of age should receive 45, 52, and 58 a second dose after 1-2 months and a third dose after 6 months) Can be administered regardless of history of abnor- mal PAP smear -

Vaccinia (Smallpox) Vaccine
December 13, 1991 / Vol. 40 / No. RR-14 Recommendations and Reports '°0 : o r Vaccinia (Smallpox) Vaccine Recommendations of the Immunization Practices Advisory Committee (ACIP) REVERT/ALUMNI LIBRARY CREIGHTOH UNIVERSITY DEPOSITORY ITEM U.S. DEPARTMENT OF HEALTH AND HUMAN SERVICES Public Health Service , Centers for Disease Control f+ National Center for Prevention Services C Atlanta, Georgia 30333 CDCV« The MMWR series of publications is published by the Epidemiology Program Office, Centers for Disease Control, Public Health Service, U.S. Department of Health and Human Services, Atlanta, Georgia 30333. SUGGESTED CITATION Centers for Disease Control. Vaccinia (smallpox) vaccine. Recommendations of the Immunization Practices Advisory Committee (ACIP). MMWR 1991;40(RR-14) [inclusive page numbers]. Centers for Disease Control.... ..William L. Roper, M.D., M.P.H. Director The material in this report was prepared for publication by: National Center for Prevention Services .Alan R. Hinman, M.D., M.P.H. Director Division of Immunization Walter A. Orenstein, M.D. Director The production of this report as an MMWR serial publication was coordinated in: Epidemiology Program Office. Stephen B. Thacker, M.D., M.Sc. Director Richard A. Goodman, M.D., M.P.H. Editor, MMWR Series Scientific Communications Program Public Health Publications Branch. Suzanne M. Hewitt Chief Karen R. Bradfield Ava W. Navin, M.A. Project Editors Morie E. Miller Editorial Assistant Use of trade names is for identification only and does not imply endorsement by the Public Health -

ACAM2000 Benefit from the Vaccine
-------------------------------CONTRAINDICATIONS------------------------------ HIGHLIGHTS OF PRESCRIBING INFORMATION • Individuals with severe immunodeficiency who are not expected to These highlights do not include all information needed to use ACAM2000 benefit from the vaccine. These individuals may include persons who safely and effectively. See full prescribing information for ACAM2000. are undergoing bone marrow transplantation or persons with primary or ACAM2000, (Smallpox (Vaccinia) Vaccine, Live) acquired immunodeficiency states who require isolation (4). Lyophilized preparation for percutaneous scarification -- ---------------------WARNINGS AND PRECAUTIONS------------------------ Initial U.S. Approval: 2007 • Myocarditis and/or pericarditis, ischemic heart disease and non-ischemic WARNING: dilated cardiomyopathy. (5.1, 5.2) See full prescribing information for complete boxed warning • Encephalitis, encephalomyelitis, encephalopathy, progressive vaccinia • Myocarditis and pericarditis (suspect cases observed at a rate of 5.7 (vaccinia necrosum), generalized vaccinia, severe vaccinial skin per 1000 primary vaccinees (95% CI: 1.9-13.3)), encephalitis, infections, erythema multiforme major (including Stevens-Johnson encephalomyelitis, encephalopathy, progressive vaccinia, generalized syndrome), eczema vaccinatum, fetal vaccinia and fetal death. (5.1) vaccinia, severe vaccinial skin infections, erythema multiforme major (including STEVENS-JOHNSON SYNDROME), eczema • Ocular vaccinia and blindness. (5.3) vaccinatum resulting in permanent