Fibrosis Accumulation in Idiopathic Pulmonary Apoptosis
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Supplemental Information to Mammadova-Bach Et Al., “Laminin Α1 Orchestrates VEGFA Functions in the Ecosystem of Colorectal Carcinogenesis”
Supplemental information to Mammadova-Bach et al., “Laminin α1 orchestrates VEGFA functions in the ecosystem of colorectal carcinogenesis” Supplemental material and methods Cloning of the villin-LMα1 vector The plasmid pBS-villin-promoter containing the 3.5 Kb of the murine villin promoter, the first non coding exon, 5.5 kb of the first intron and 15 nucleotides of the second villin exon, was generated by S. Robine (Institut Curie, Paris, France). The EcoRI site in the multi cloning site was destroyed by fill in ligation with T4 polymerase according to the manufacturer`s instructions (New England Biolabs, Ozyme, Saint Quentin en Yvelines, France). Site directed mutagenesis (GeneEditor in vitro Site-Directed Mutagenesis system, Promega, Charbonnières-les-Bains, France) was then used to introduce a BsiWI site before the start codon of the villin coding sequence using the 5’ phosphorylated primer: 5’CCTTCTCCTCTAGGCTCGCGTACGATGACGTCGGACTTGCGG3’. A double strand annealed oligonucleotide, 5’GGCCGGACGCGTGAATTCGTCGACGC3’ and 5’GGCCGCGTCGACGAATTCACGC GTCC3’ containing restriction site for MluI, EcoRI and SalI were inserted in the NotI site (present in the multi cloning site), generating the plasmid pBS-villin-promoter-MES. The SV40 polyA region of the pEGFP plasmid (Clontech, Ozyme, Saint Quentin Yvelines, France) was amplified by PCR using primers 5’GGCGCCTCTAGATCATAATCAGCCATA3’ and 5’GGCGCCCTTAAGATACATTGATGAGTT3’ before subcloning into the pGEMTeasy vector (Promega, Charbonnières-les-Bains, France). After EcoRI digestion, the SV40 polyA fragment was purified with the NucleoSpin Extract II kit (Machery-Nagel, Hoerdt, France) and then subcloned into the EcoRI site of the plasmid pBS-villin-promoter-MES. Site directed mutagenesis was used to introduce a BsiWI site (5’ phosphorylated AGCGCAGGGAGCGGCGGCCGTACGATGCGCGGCAGCGGCACG3’) before the initiation codon and a MluI site (5’ phosphorylated 1 CCCGGGCCTGAGCCCTAAACGCGTGCCAGCCTCTGCCCTTGG3’) after the stop codon in the full length cDNA coding for the mouse LMα1 in the pCIS vector (kindly provided by P. -

Gene Symbol Gene Description ACVR1B Activin a Receptor, Type IB
Table S1. Kinase clones included in human kinase cDNA library for yeast two-hybrid screening Gene Symbol Gene Description ACVR1B activin A receptor, type IB ADCK2 aarF domain containing kinase 2 ADCK4 aarF domain containing kinase 4 AGK multiple substrate lipid kinase;MULK AK1 adenylate kinase 1 AK3 adenylate kinase 3 like 1 AK3L1 adenylate kinase 3 ALDH18A1 aldehyde dehydrogenase 18 family, member A1;ALDH18A1 ALK anaplastic lymphoma kinase (Ki-1) ALPK1 alpha-kinase 1 ALPK2 alpha-kinase 2 AMHR2 anti-Mullerian hormone receptor, type II ARAF v-raf murine sarcoma 3611 viral oncogene homolog 1 ARSG arylsulfatase G;ARSG AURKB aurora kinase B AURKC aurora kinase C BCKDK branched chain alpha-ketoacid dehydrogenase kinase BMPR1A bone morphogenetic protein receptor, type IA BMPR2 bone morphogenetic protein receptor, type II (serine/threonine kinase) BRAF v-raf murine sarcoma viral oncogene homolog B1 BRD3 bromodomain containing 3 BRD4 bromodomain containing 4 BTK Bruton agammaglobulinemia tyrosine kinase BUB1 BUB1 budding uninhibited by benzimidazoles 1 homolog (yeast) BUB1B BUB1 budding uninhibited by benzimidazoles 1 homolog beta (yeast) C9orf98 chromosome 9 open reading frame 98;C9orf98 CABC1 chaperone, ABC1 activity of bc1 complex like (S. pombe) CALM1 calmodulin 1 (phosphorylase kinase, delta) CALM2 calmodulin 2 (phosphorylase kinase, delta) CALM3 calmodulin 3 (phosphorylase kinase, delta) CAMK1 calcium/calmodulin-dependent protein kinase I CAMK2A calcium/calmodulin-dependent protein kinase (CaM kinase) II alpha CAMK2B calcium/calmodulin-dependent -

Tumour-Agnostic Therapy for Pancreatic Cancer and Biliary Tract Cancer
diagnostics Review Tumour-Agnostic Therapy for Pancreatic Cancer and Biliary Tract Cancer Shunsuke Kato Department of Clinical Oncology, Juntendo University Graduate School of Medicine, 2-1-1, Hongo, Bunkyo-ku, Tokyo 113-8421, Japan; [email protected]; Tel.: +81-3-5802-1543 Abstract: The prognosis of patients with solid tumours has remarkably improved with the develop- ment of molecular-targeted drugs and immune checkpoint inhibitors. However, the improvements in the prognosis of pancreatic cancer and biliary tract cancer is delayed compared to other carcinomas, and the 5-year survival rates of distal-stage disease are approximately 10 and 20%, respectively. How- ever, a comprehensive analysis of tumour cells using The Cancer Genome Atlas (TCGA) project has led to the identification of various driver mutations. Evidently, few mutations exist across organs, and basket trials targeting driver mutations regardless of the primary organ are being actively conducted. Such basket trials not only focus on the gate keeper-type oncogene mutations, such as HER2 and BRAF, but also focus on the caretaker-type tumour suppressor genes, such as BRCA1/2, mismatch repair-related genes, which cause hereditary cancer syndrome. As oncogene panel testing is a vital approach in routine practice, clinicians should devise a strategy for improved understanding of the cancer genome. Here, the gene mutation profiles of pancreatic cancer and biliary tract cancer have been outlined and the current status of tumour-agnostic therapy in these cancers has been reported. Keywords: pancreatic cancer; biliary tract cancer; targeted therapy; solid tumours; driver mutations; agonist therapy Citation: Kato, S. Tumour-Agnostic Therapy for Pancreatic Cancer and 1. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

In Vivo Dual RNA-Seq Analysis Reveals the Basis for Differential Tissue Tropism of Clinical Isolates of Streptococcus Pneumoniae
In Vivo Dual RNA-Seq Analysis Reveals the Basis for Differential Tissue Tropism of Clinical Isolates of Streptococcus pneumoniae Vikrant Minhas,1,4 Rieza Aprianto,2,4 Lauren J. McAllister,1 Hui Wang,1 Shannon C. David,1 Kimberley T. McLean,1 Iain Comerford,3 Shaun R. McColl,3 James C. Paton,1,5,6,* Jan-Willem Veening,2,5 and Claudia Trappetti,1,5 Supplementary Information Supplementary Table 1. Pneumococcal differential gene expression in the lungs 6 h post-infection, 9-47-Ear vs 9-47M. Genes with fold change (FC) greater than 2 and p < 0.05 are shown. FC values highlighted in blue = upregulated in 9-47-Ear, while values highlighted in red = upregulated in 9- 47M. Locus tag in 9-47- Product padj FC Ear Sp947_chr_00844 Sialidase B 3.08E-10 313.9807 Sp947_chr_02077 hypothetical protein 4.46E-10 306.9412 Sp947_chr_00842 Sodium/glucose cotransporter 2.22E-09 243.4822 Sp947_chr_00841 N-acetylneuraminate lyase 4.53E-09 227.7963 scyllo-inositol 2-dehydrogenase Sp947_chr_00845 (NAD(+)) 4.36E-09 221.051 Sp947_chr_00848 hypothetical protein 1.19E-08 202.7867 V-type sodium ATPase catalytic subunit Sp947_chr_00853 A 1.29E-06 100.5411 Sp947_chr_00846 Beta-glucoside kinase 3.42E-06 98.18951 Sp947_chr_00855 V-type sodium ATPase subunit D 8.34E-06 85.94879 Sp947_chr_00851 V-type sodium ATPase subunit C 2.50E-05 72.46612 Sp947_chr_00843 hypothetical protein 2.17E-05 65.97758 Sp947_chr_00839 HTH-type transcriptional regulator RpiR 3.09E-05 61.28171 Sp947_chr_00854 V-type sodium ATPase subunit B 1.32E-06 50.86992 Sp947_chr_00120 hypothetical protein 3.00E-04 -

Inflammation-Dependent IL18 Signaling Restricts Hepatocellular Carcinoma Growth by Enhancing the Accumulation and Activity of Tumor-Infiltrating Lymphocytes
Published OnlineFirst February 18, 2016; DOI: 10.1158/0008-5472.CAN-15-1548 Cancer Tumor and Stem Cell Biology Research Inflammation-Dependent IL18 Signaling Restricts Hepatocellular Carcinoma Growth by Enhancing the Accumulation and Activity of Tumor- Infiltrating Lymphocytes Geoffrey J. Markowitz1, Pengyuan Yang1,2,3, Jing Fu3, Gregory A. Michelotti4, Rui Chen1, Jianhua Sui5, Bin Yang2, Wen-Hao Qin3, Zheng Zhang6, Fu-Sheng Wang6, Anna Mae Diehl4, Qi-Jing Li7, Hongyang Wang3, and Xiao-Fan Wang1 Abstract Chronic inflammation in liver tissue is an underlying cause of IL18R1 deletion increased tumor burden. Mechanistically, we hepatocellular carcinoma. High levels of inflammatory cytokine foundthatIL18exertedinflammation-dependent tumor-sup- IL18 in the circulation of patients with hepatocellular carcinoma pressive effects largely by promoting the differentiation, activ- correlates with poor prognosis. However, conflicting results have ity, and survival of tumor-infiltrating T cells. Finally, differences been reported for IL18 in hepatocellular carcinoma development in the expression of IL18 in tumor tissue versus nontumor and progression. In this study, we used tissue specimens from tissueweremorepredictiveofpatientoutcomethanoverall hepatocellular carcinoma patients and clinically relevant mouse tissue expression. Taken together, our findings resolve a long- models of hepatocellular carcinoma to evaluate IL18 expression standing contradiction regarding a tumor-suppressive role for and function. In a mouse model of liver fibrosis that recapitulates IL18 in established hepatocellular carcinoma and provide a a tumor-promoting microenvironment, global deletion of the mechanistic explanation for the complex relationship between IL18 receptor IL18R1 enhanced tumor growth and burden. Sim- its expression pattern and hepatocellular carcinoma prognosis. ilarly, in a carcinogen-induced model of liver tumorigenesis, Cancer Res; 76(8); 1–12. -

PDF Download
MEK-7 Polyclonal Antibody Catalog No : YT2725 Reactivity : Human,Mouse,Rat Applications : WB,ELISA Gene Name : MAP2K7 Protein Name : Dual specificity mitogen-activated protein kinase kinase 7 Human Gene Id : 5609 Human Swiss Prot O14733 No : Mouse Gene Id : 26400 Mouse Swiss Prot Q8CE90 No : Rat Gene Id : 363855 Rat Swiss Prot No : Q4KSH7 Immunogen : The antiserum was produced against synthesized peptide derived from human MAP2K7. AA range:241-290 Specificity : MEK-7 Polyclonal Antibody detects endogenous levels of MEK-7 protein. Formulation : Liquid in PBS containing 50% glycerol, 0.5% BSA and 0.02% sodium azide. Source : Rabbit Dilution : Western Blot: 1/500 - 1/2000. ELISA: 1/20000. Not yet tested in other applications. Purification : The antibody was affinity-purified from rabbit antiserum by affinity- chromatography using epitope-specific immunogen. Concentration : 1 mg/ml 1 / 3 Storage Stability : -20°C/1 year Molecularweight : 47485 Observed Band : 47 Cell Pathway : MAPK_ERK_Growth,MAPK_G_Protein,ErbB_HER,Toll_Like,T_Cell_Receptor, Fc epsilon RI,Neurotrophin,GnRH, Background : mitogen-activated protein kinase kinase 7(MAP2K7) Homo sapiens The protein encoded by this gene is a dual specificity protein kinase that belongs to the MAP kinase kinase family. This kinase specifically activates MAPK8/JNK1 and MAPK9/JNK2, and this kinase itself is phosphorylated and activated by MAP kinase kinase kinases including MAP3K1/MEKK1, MAP3K2/MEKK2,MAP3K3/MEKK5, and MAP4K2/GCK. This kinase is involved in the signal transduction mediating the cell -

Application of a MYC Degradation
SCIENCE SIGNALING | RESEARCH ARTICLE CANCER Copyright © 2019 The Authors, some rights reserved; Application of a MYC degradation screen identifies exclusive licensee American Association sensitivity to CDK9 inhibitors in KRAS-mutant for the Advancement of Science. No claim pancreatic cancer to original U.S. Devon R. Blake1, Angelina V. Vaseva2, Richard G. Hodge2, McKenzie P. Kline3, Thomas S. K. Gilbert1,4, Government Works Vikas Tyagi5, Daowei Huang5, Gabrielle C. Whiten5, Jacob E. Larson5, Xiaodong Wang2,5, Kenneth H. Pearce5, Laura E. Herring1,4, Lee M. Graves1,2,4, Stephen V. Frye2,5, Michael J. Emanuele1,2, Adrienne D. Cox1,2,6, Channing J. Der1,2* Stabilization of the MYC oncoprotein by KRAS signaling critically promotes the growth of pancreatic ductal adeno- carcinoma (PDAC). Thus, understanding how MYC protein stability is regulated may lead to effective therapies. Here, we used a previously developed, flow cytometry–based assay that screened a library of >800 protein kinase inhibitors and identified compounds that promoted either the stability or degradation of MYC in a KRAS-mutant PDAC cell line. We validated compounds that stabilized or destabilized MYC and then focused on one compound, Downloaded from UNC10112785, that induced the substantial loss of MYC protein in both two-dimensional (2D) and 3D cell cultures. We determined that this compound is a potent CDK9 inhibitor with a previously uncharacterized scaffold, caused MYC loss through both transcriptional and posttranslational mechanisms, and suppresses PDAC anchorage- dependent and anchorage-independent growth. We discovered that CDK9 enhanced MYC protein stability 62 through a previously unknown, KRAS-independent mechanism involving direct phosphorylation of MYC at Ser . -

Serum Albumin OS=Homo Sapiens
Protein Name Cluster of Glial fibrillary acidic protein OS=Homo sapiens GN=GFAP PE=1 SV=1 (P14136) Serum albumin OS=Homo sapiens GN=ALB PE=1 SV=2 Cluster of Isoform 3 of Plectin OS=Homo sapiens GN=PLEC (Q15149-3) Cluster of Hemoglobin subunit beta OS=Homo sapiens GN=HBB PE=1 SV=2 (P68871) Vimentin OS=Homo sapiens GN=VIM PE=1 SV=4 Cluster of Tubulin beta-3 chain OS=Homo sapiens GN=TUBB3 PE=1 SV=2 (Q13509) Cluster of Actin, cytoplasmic 1 OS=Homo sapiens GN=ACTB PE=1 SV=1 (P60709) Cluster of Tubulin alpha-1B chain OS=Homo sapiens GN=TUBA1B PE=1 SV=1 (P68363) Cluster of Isoform 2 of Spectrin alpha chain, non-erythrocytic 1 OS=Homo sapiens GN=SPTAN1 (Q13813-2) Hemoglobin subunit alpha OS=Homo sapiens GN=HBA1 PE=1 SV=2 Cluster of Spectrin beta chain, non-erythrocytic 1 OS=Homo sapiens GN=SPTBN1 PE=1 SV=2 (Q01082) Cluster of Pyruvate kinase isozymes M1/M2 OS=Homo sapiens GN=PKM PE=1 SV=4 (P14618) Glyceraldehyde-3-phosphate dehydrogenase OS=Homo sapiens GN=GAPDH PE=1 SV=3 Clathrin heavy chain 1 OS=Homo sapiens GN=CLTC PE=1 SV=5 Filamin-A OS=Homo sapiens GN=FLNA PE=1 SV=4 Cytoplasmic dynein 1 heavy chain 1 OS=Homo sapiens GN=DYNC1H1 PE=1 SV=5 Cluster of ATPase, Na+/K+ transporting, alpha 2 (+) polypeptide OS=Homo sapiens GN=ATP1A2 PE=3 SV=1 (B1AKY9) Fibrinogen beta chain OS=Homo sapiens GN=FGB PE=1 SV=2 Fibrinogen alpha chain OS=Homo sapiens GN=FGA PE=1 SV=2 Dihydropyrimidinase-related protein 2 OS=Homo sapiens GN=DPYSL2 PE=1 SV=1 Cluster of Alpha-actinin-1 OS=Homo sapiens GN=ACTN1 PE=1 SV=2 (P12814) 60 kDa heat shock protein, mitochondrial OS=Homo -

WO 2019/079361 Al 25 April 2019 (25.04.2019) W 1P O PCT
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization I International Bureau (10) International Publication Number (43) International Publication Date WO 2019/079361 Al 25 April 2019 (25.04.2019) W 1P O PCT (51) International Patent Classification: CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, C12Q 1/68 (2018.01) A61P 31/18 (2006.01) DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, C12Q 1/70 (2006.01) HR, HU, ID, IL, IN, IR, IS, JO, JP, KE, KG, KH, KN, KP, KR, KW, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, (21) International Application Number: MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, PCT/US2018/056167 OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, (22) International Filing Date: SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, 16 October 2018 (16. 10.2018) TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (25) Filing Language: English (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (26) Publication Language: English GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, ST, SZ, TZ, (30) Priority Data: UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, 62/573,025 16 October 2017 (16. 10.2017) US TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, EE, ES, FI, FR, GB, GR, HR, HU, ΓΕ , IS, IT, LT, LU, LV, (71) Applicant: MASSACHUSETTS INSTITUTE OF MC, MK, MT, NL, NO, PL, PT, RO, RS, SE, SI, SK, SM, TECHNOLOGY [US/US]; 77 Massachusetts Avenue, TR), OAPI (BF, BJ, CF, CG, CI, CM, GA, GN, GQ, GW, Cambridge, Massachusetts 02139 (US). -
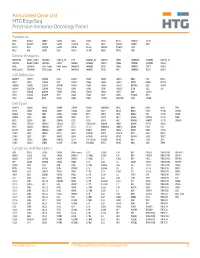
Annotated Gene List HTG Edgeseq Precision Immuno-Oncology Panel
Annotated Gene List HTG EdgeSeq Precision Immuno-Oncology Panel Apoptosis APAF1 BCL2L1 CARD11 CASP4 CD5L FADD KSR2 OPTN SAMD12 TCF19 BAX BCL2L11 CASP1 CASP5 CORO1A FAS LRG1 PLA2G6 SAMD9 XAF1 BCL10 BCL6 CASP10 CASP8 DAPK2 FASLG MECOM PYCARD SPOP BCL2 BID CASP3 CAV1 DAPL1 GLIPR1 MELK RIPK2 TBK1 Cancer Antigens ANKRD30A BAGE2_BAGE3 CEACAM6 CTAG1A_1B LIPE MAGEA3_A6 MAGEC2 PAGE3 SPANXACD SPANXN4 XAGE1B_1E ARMCX6 BAGE4_BAGE5 CEACAM8 CTAG2 MAGEA1 MAGEA4 MTFR2 PAGE4 SPANXB1 SPANXN5 XAGE2 BAGE CEACAM1 CT45_family GAGE_family MAGEA10 MAGEB2 PAGE1 PAGE5 SPANXN1 SYCP1 XAGE3 BAGE_family CEACAM5 CT47_family HPN MAGEA12 MAGEC1 PAGE2 PBK SPANXN3 TEX14 XAGE5 Cell Adhesion ADAM17 CDH15 CLEC5A DSG3 ICAM2 ITGA5 ITGB2 LAMC3 MBL2 PVR UPK2 ADD2 CDH5 CLEC6A DST ICAM3 ITGA6 ITGB3 LAMP1 MTDH RRAS2 UPK3A ADGRE5 CLDN3 CLEC7A EPCAM ICAM4 ITGAE ITGB4 LGALS1 NECTIN2 SELE VCAM1 ALCAM CLEC12A CLEC9A FBLN1 ITGA1 ITGAL ITGB7 LGALS3 OCLN SELL ZYX CD63 CLEC2B DIAPH3 FXYD5 ITGA2 ITGAM ITLN2 LYVE1 OLR1 SELPLG CD99 CLEC4A DLGAP5 IBSP ITGA3 ITGAX JAML M6PR PECAM1 THY1 CDH1 CLEC4C DSC3 ICAM1 ITGA4 ITGB1 L1CAM MADCAM1 PKP1 UNC5D Cell Cycle ANAPC1 CCND3 CDCA5 CENPH CNNM1 ESCO2 HORMAD2 KIF2C MELK ORC6 SKA3 TPX2 ASPM CCNE1 CDCA8 CENPI CNTLN ESPL1 IKZF1 KIF4A MND1 PATZ1 SP100 TRIP13 AURKA CCNE2 CDK1 CENPL CNTLN ETS1 IKZF2 KIF5C MYBL2 PIF1 SP110 TROAP AURKB CCNF CDK4 CENPU DBF4 ETS2 IKZF3 KIFC1 NCAPG PIMREG SPC24 TUBB BEX1 CDC20 CDK6 CENPW E2F2 EZH2 IKZF4 KNL1 NCAPG2 PKMYT1 SPC25 ZWILCH BEX2 CDC25A CDKN1A CEP250 E2F7 GADD45GIP1 KDM5B LMNA NCAPH POC1A SPDL1 BUB1 CDC25C CDKN1B CEP55 ECT2 -

Spatial Protein Interaction Networks of the Intrinsically Disordered Transcription Factor C(%3$
Spatial protein interaction networks of the intrinsically disordered transcription factor C(%3$ Dissertation zur Erlangung des akademischen Grades Doctor rerum naturalium (Dr. rer. nat.) im Fach Biologie/Molekularbiologie eingereicht an der Lebenswissenschaftlichen Fakultät der Humboldt-Universität zu Berlin Von Evelyn Ramberger, M.Sc. Präsidentin der Humboldt-Universität zu Berlin Prof. Dr.-Ing.Dr. Sabine Kunst Dekan der Lebenswissenschaftlichen Fakultät der Humboldt-Universität zu Berlin Prof. Dr. Bernhard Grimm Gutachter: 1. Prof. Dr. Achim Leutz 2. Prof. Dr. Matthias Selbach 3. Prof. Dr. Gunnar Dittmar Tag der mündlichen Prüfung: 12.8.2020 For T. Table of Contents Selbstständigkeitserklärung ....................................................................................1 List of Figures ............................................................................................................2 List of Tables ..............................................................................................................3 Abbreviations .............................................................................................................4 Zusammenfassung ....................................................................................................6 Summary ....................................................................................................................7 1. Introduction ............................................................................................................8 1.1. Disordered proteins