Induction of Apoptosis Increases Expression of Non-Canonical WNT Genes in Myeloid Leukemia Cell Lines
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

A High-Fat/High-Sucrose Diet Induces WNT4 Expression in Mouse Pancreatic Β-Cells
This is “Advance Publication Article” Kurume Medical Journal, 65, 00-00, 2018 Original Article A High-Fat/High-Sucrose Diet Induces WNT4 Expression in Mouse Pancreatic β-cells YAYOI KURITA, TSUYOSHI OHKI, ERI SOEJIMA, XIAOHONG YUAN, SATOMI KAKINO, NOBUHIKO WADA, TOSHIHIKO HASHINAGA, HITOMI NAKAYAMA, JUNICHI TANI, YUJI TAJIRI, YUJI HIROMATSU, KENTARO YAMADA* AND MASATOSHI NOMURA Division of Endocrinology and Metabolism, Department of Internal Medicine, Kurume University School of Medicine, Kurume 830-0011, Japan, *Diabetes Center of Asakura Medical Association Hospital, Asakura 838-0069, Japan Received 12 February 2018, accepted 1 May 2018 J-STAGE advance publication 11 March 2019 Edited by MAKOTO TAKANO Summary: Aims/Introduction: Several lines of evidence suggest that dysregulation of the WNT signaling pathway is involved in the pathogenesis of type 2 diabetes. This study was performed to elucidate the effects of a high-fat/high-sucrose (HF/HS) diet on pancreatic islet functions in relation to modulation of WNT ligand expression in β-cells. Materials and Methods: Mice were fed either standard mouse chow or a HF/HS diet from 8 weeks of age. At 20 weeks of age, intraperitoneal glucose tolerance tests were performed in both groups of mice, followed by euthanasia and isolation of pancreatic islets. WNT-related gene expression in islets and MIN6 cells was measured by quantitative real-time RT-PCR. To explore the direct effects of WNT signals on pancreatic β-cells, MIN6 cells were exposed to recombinant mouse WNT4 protein (rmWNT4) for 48 h, and glucose-induced insulin secre- tion was measured. Furthermore, Wnt4 siRNAs were transfected into MIN6 cells, and cell viability and insulin secretion were measured in control and Wnt4 siRNA-transfected MIN6 cells. -

WNT10A Gene Wnt Family Member 10A
WNT10A gene Wnt family member 10A Normal Function The WNT10A gene is part of a large family of WNT genes, which play critical roles in development starting before birth. These genes provide instructions for making proteins that participate in chemical signaling pathways in the body. Wnt signaling controls the activity of certain genes and regulates the interactions between cells during embryonic development. The protein produced from the WNT10A gene plays a role in the development of many parts of the body. It appears to be essential for the formation of tissues that arise from an embryonic cell layer called the ectoderm. These tissues include the skin, hair, nails, teeth, and sweat glands. Researchers believe that the WNT10A protein is particularly important for the formation and shaping of both baby (primary) teeth and adult ( permanent) teeth. Health Conditions Related to Genetic Changes Hypohidrotic ectodermal dysplasia Several mutations in the WNT10A gene have been found to cause hypohidrotic ectodermal dysplasia, the most common form of ectodermal dysplasia. Starting before birth, ectodermal dysplasias result in the abnormal development of the skin, hair, nails, teeth, and sweat glands. Hypohidrotic ectodermal dysplasia is characterized by a reduced ability to sweat (hypohidrosis), sparse scalp and body hair (hypotrichosis), and several missing teeth (hypodontia) or teeth that are malformed. WNT10A gene mutations account for about 5 percent of all cases of hypohidrotic ectodermal dysplasia. Most of the WNT10A gene mutations associated with hypohidrotic ectodermal dysplasia change single protein building blocks (amino acids) in the WNT10A protein, which impairs its function. The resulting shortage of functional WNT10A protein disrupts Wnt signaling during the development of ectodermal tissues, particularly the teeth. -

The Sec14-Like Phosphatidylinositol Transfer Proteins Sec14l3/SEC14L2
RESEARCH ARTICLE The Sec14-like phosphatidylinositol transfer proteins Sec14l3/SEC14L2 act as GTPase proteins to mediate Wnt/Ca2+ signaling Bo Gong, Weimin Shen, Wanghua Xiao, Yaping Meng, Anming Meng*, Shunji Jia* State Key Laboratory of Membrane Biology, Tsinghua-Peking Center for Life Sciences, School of Life Sciences, Tsinghua University, Beijing, China Abstract The non-canonical Wnt/Ca2+ signaling pathway plays important roles in embryonic development, tissue formation and diseases. However, it is unclear how the Wnt ligand-stimulated, G protein-coupled receptor Frizzled activates phospholipases for calcium release. Here, we report that the zebrafish/human phosphatidylinositol transfer protein Sec14l3/SEC14L2 act as GTPase proteins to transduce Wnt signals from Frizzled to phospholipase C (PLC). Depletion of sec14l3 attenuates Wnt/Ca2+ responsive activity and causes convergent and extension (CE) defects in zebrafish embryos. Biochemical analyses in mammalian cells indicate that Sec14l3-GDP forms complex with Frizzled and Dishevelled; Wnt ligand binding of Frizzled induces translocation of Sec14l3 to the plasma membrane; and then Sec14l3-GTP binds to and activates phospholipase Cd4a (Plcd4a); subsequently, Plcd4a initiates phosphatidylinositol-4,5-bisphosphate (PIP2) signaling, ultimately stimulating calcium release. Furthermore, Plcd4a can act as a GTPase-activating protein to accelerate the hydrolysis of Sec14l3-bound GTP to GDP. Our data provide a new insight into GTPase protein-coupled Wnt/Ca2+ signaling transduction. DOI: 10.7554/eLife.26362.001 *For correspondence: mengam@ mail.tsinghua.edu.cn (AM); jiasj@ mail.tsinghua.edu.cn (SJ) Competing interests: The Introduction authors declare that no Wnt ligands, a large family of secreted lipoglycoproteins, control a large number of developmental competing interests exist. -

Activation of Thewnt–Яcatenin Pathway in a Cell Population on The
The Journal of Neuroscience, September 5, 2007 • 27(36):9757–9768 • 9757 Development/Plasticity/Repair Activation of the Wnt–Catenin Pathway in a Cell Population on the Surface of the Forebrain Is Essential for the Establishment of Olfactory Axon Connections Ambra A. Zaghetto,1 Sara Paina,1 Stefano Mantero,1 Natalia Platonova,1 Paolo Peretto,2 Serena Bovetti,2,3 Adam Puche,3 Stefano Piccolo,4 and Giorgio R. Merlo1 1Dulbecco Telethon Institute-Consiglio Nazionale delle Ricerche Institute for Biomedical Technologies Milano, 20090 Segrate, Italy, 2Department of Animal and Human Biology, University of Torino, 10123 Torino, Italy, 3Department of Anatomy and Neurobiology, School of Medicine, University of Maryland, Baltimore, Maryland 21201, and 4Department of Histology, Microbiology, and Medical Biotechnologies, School of Medicine, University of Padova, 35122 Padova, Italy A variety of signals governing early extension, guidance, and connectivity of olfactory receptor neuron (ORN) axons has been identified; however, little is known about axon–mesoderm and forebrain (FB)–mesoderm signals. Using Wnt–catenin reporter mice, we identify a novel Wnt-responsive resident cell population, located in a Frizzled7 expression domain at the surface of the embryonic FB, along the trajectory of incoming ORN axons. Organotypic slice cultures that recapitulate olfactory-associated Wnt–catenin activation show that the catenin response depends on a placode-derived signal(s). Likewise, in Dlx5Ϫ/Ϫ embryos, in which the primary connections fail to form, Wnt–catenin response on the surface of the FB is strongly reduced. The olfactory placode expresses a number of catenin- activating Wnt genes, and the Frizzled7 receptor transduces the “canonical” Wnt signal; using Wnt expression plasmids we show that Wnt5a and Wnt7b are sufficient to rescue catenin activation in the absence of incoming axons. -

Towards an Integrated View of Wnt Signaling in Development Renée Van Amerongen and Roel Nusse*
HYPOTHESIS 3205 Development 136, 3205-3214 (2009) doi:10.1242/dev.033910 Towards an integrated view of Wnt signaling in development Renée van Amerongen and Roel Nusse* Wnt signaling is crucial for embryonic development in all animal Notably, components at virtually every level of the Wnt signal species studied to date. The interaction between Wnt proteins transduction cascade have been shown to affect both β-catenin- and cell surface receptors can result in a variety of intracellular dependent and -independent responses, depending on the cellular responses. A key remaining question is how these specific context. As we discuss below, this holds true for the Wnt proteins responses take shape in the context of a complex, multicellular themselves, as well as for their receptors and some intracellular organism. Recent studies suggest that we have to revise some of messengers. Rather than concluding that these proteins are shared our most basic ideas about Wnt signal transduction. Rather than between pathways, we instead propose that it is the total net thinking about Wnt signaling in terms of distinct, linear, cellular balance of signals that ultimately determines the response of the signaling pathways, we propose a novel view that considers the receiving cell. In the context of an intact and developing integration of multiple, often simultaneous, inputs at the level organism, cells receive multiple, dynamic, often simultaneous and of both Wnt-receptor binding and the downstream, sometimes even conflicting inputs, all of which are integrated to intracellular response. elicit the appropriate cell behavior in response. As such, the different signaling pathways might thus be more intimately Introduction intertwined than previously envisioned. -

Role of DNA Methylation in Adipogenesis
Georgia State University ScholarWorks @ Georgia State University Biology Theses Department of Biology Summer 8-12-2014 Role of DNA Methylation in Adipogenesis Yii-Shyuan Chen Follow this and additional works at: https://scholarworks.gsu.edu/biology_theses Recommended Citation Chen, Yii-Shyuan, "Role of DNA Methylation in Adipogenesis." Thesis, Georgia State University, 2014. https://scholarworks.gsu.edu/biology_theses/57 This Thesis is brought to you for free and open access by the Department of Biology at ScholarWorks @ Georgia State University. It has been accepted for inclusion in Biology Theses by an authorized administrator of ScholarWorks @ Georgia State University. For more information, please contact [email protected]. ROLE OF DNA METHYLATION IN ADIPOGENESIS by YII-SHYUAN CHEN Under the Direction of Bingzhong Xue ABSTRACT The increase in the prevalence of obesity and obesity-related diseases has caused greater attention to be placed on the molecular mechanisms controlling adipogenesis. In this study, we studied the role of 5-aza-2'-deoxycytidine (5-Aza-dC), an inhibitor of DNA methylation, on adipocyte differentiation. We found that inhibiting DNA methylation by 5-Aza-dC significantly inhibited adipocyte differentiation whereas promoting osteoblastogenesis. Wnt10a was up- regulated by 5-Aza-dC treatment and it was suggested that Wnt10a might play a vital role in suppressing adipogenesis and promoting osteoblastogenesis by inhibiting DNA methylation. In 3T3-L1 cells, Wnt signaling inhibitor IWP-2 was found to reverse the inhibitory effect of 5-Aza- dC on Adipocyte differentiation, whereas in mesenchymal stem cell line, ST2 cells, IWP-2 treatment reversed the effect of 5-Aza-dC on promoting osteoblastogenesis. -

Control of Wnt5b Secretion by Wntless Modulates Chondrogenic Cell Proliferation Through Fine-Tuning Fgf3 Expression Bo-Tsung Wu1,2, Shih-Hsien Wen1,2, Sheng-Ping L
© 2015. Published by The Company of Biologists Ltd | Journal of Cell Science (2015) 128, 2328-2339 doi:10.1242/jcs.167403 RESEARCH ARTICLE Control of Wnt5b secretion by Wntless modulates chondrogenic cell proliferation through fine-tuning fgf3 expression Bo-Tsung Wu1,2, Shih-Hsien Wen1,2, Sheng-Ping L. Hwang3, Chang-Jen Huang1,2 and Yung-Shu Kuan1,2,4,* ABSTRACT activities to achieve the proper proliferation, differentiation or Wnts and Fgfs regulate various tissues development in migration responses is still relatively limited. β vertebrates. However, how regional Wnt or Fgf activities are In vertebrates, the -catenin-mediated canonical and the non- established and how they interact in any given developmental canonical Wnt signaling pathways have both been shown to be event is elusive. Here, we investigated the Wnt-mediated involved in the processes of craniofacial skeleton formation. In craniofacial cartilage development in zebrafish and found that mice, previous observations have indicated that Wnts can either fgf3 expression in the pharyngeal pouches is differentially reduced stimulate chondrogenesis by promoting survival and differentiation along the anteroposterior axis in wnt5b mutants and wntless (wls) of migrating neural crest cells (NCCs), or inhibit chondrogenesis by morphants, but its expression is normal in wnt9a and wnt11 repressing BMP2-induced chondrocyte gene expression, depending morphants. Introducing fgf3 mRNAs rescued the cartilage defects on the developmental stage and the local tissue context (Brault et al., in Wnt5b- and Wls-deficient larvae. In wls morphants, endogenous 2001; Liu et al., 2008; Reinhold et al., 2006; Yang et al., 2003). In Wls expression is not detectable but maternally deposited Wls is zebrafish, wnt4a and wnt11r have been shown to regulate the present in eggs, which might account for the lack of axis defects in formation of pharyngeal pouches, whereas wnt5b, wnt9a and wnt11 wls morphants. -

Wnt4/B2catenin Signaling in Medullary Kidney Myofibroblasts
BASIC RESEARCH www.jasn.org Wnt4/b2Catenin Signaling in Medullary Kidney Myofibroblasts † †‡ | Derek P. DiRocco,* Akio Kobayashi,* Makoto M. Taketo,§ Andrew P. McMahon, and †‡ Benjamin D. Humphreys* *Renal Division, Brigham and Women’s Hospital, Boston, Massachusetts; †Harvard Medical School, Boston, Massachusetts; ‡Harvard Stem Cell Institute, Cambridge, Massachusetts; §Department of Pharmacology, Graduate School of Medicine, Kyoto University, Yoshida-Konoé-cho, Sakyo, Kyoto, Japan; and |Department of Stem Cell Biology and Regenerative Medicine, Eli and Edythe Broad-CIRM Center for Regenerative Medicine and Stem Cell Research, Keck School of Medicine of the University of Southern California, Los Angeles, California ABSTRACT Injury to the adult kidney induces a number of developmental genes thought to regulate repair, including Wnt4. During kidney development, early nephron precursors and medullary stroma both express Wnt4, where it regulates epithelialization and controls smooth muscle fate, respectively. Expression patterns and roles for Wnt4 in the adult kidney, however, remain unclear. In this study, we used reporters, lineage analysis, and conditional knockout or activation of the Wnt/b-catenin pathway to investigate Wnt4 in the adult kidney. Proliferating, medullary, interstitial myofibroblasts strongly expressed Wnt4 during renal fibrosis, whereas tubule epithelia, except for the collecting duct, did not. Exogenous Wnt4 drove myofi- broblast differentiation of a pericyte-like cell line, suggesting that Wnt4 might regulate pericyte-to-myo- fibroblast transition through autocrine signaling. However, conditional deletion of Wnt4 in interstitial cells did not reduce myofibroblast proliferation, cell number, or myofibroblast gene expression during fibrosis. Because the injured kidney expresses multiple Wnt ligands that might compensate for the absence of Wnt4, we generated a mouse model with constitutive activation of canonical Wnt/b-catenin signaling in interstitial pericytes and fibroblasts. -

A Canonical to Non-Canonical Wnt Signalling Switch in Haematopoietic Stem-Cell Ageing
LETTER doi:10.1038/nature12631 A canonical to non-canonical Wnt signalling switch in haematopoietic stem-cell ageing Maria Carolina Florian1, Kalpana J. Nattamai2, Karin Do¨rr1, Gina Marka1, Bettina U¨berle1, Virag Vas1, Christina Eckl3, Immanuel Andra¨4, Matthias Schiemann4, Robert A. J. Oostendorp3, Karin Scharffetter-Kochanek1, Hans Armin Kestler5, Yi Zheng2 & Hartmut Geiger1,2 Many organs with a high cell turnover (for example, skin, intestine LT-HSCs presented with a reduced level and primarily cytoplasmic loca- and blood) are composed of short-lived cells that require continuous lization of b-catenin (Fig. 1d, e and Supplementary Video 3). Reduced replenishment by somatic stem cells1,2. Ageing results in the inability levels of b-catenin upon ageing were specific to the LT-HSC compart- of these tissues to maintain homeostasis and it is believed that somatic ment, as more differentiated LKs (Lin2c-Kit1Sca-12 cells), LSKs stem-cell ageing is one underlying cause of tissue attrition with age (Lin2Sca-11c-Kit1 cells), lymphoid-primed multipotent progenitors or age-related diseases. Ageing of haematopoietic stem cells (HSCs) (LMPPs; Lin2c-Kit1Sca-12CD341Flk21 cells) and short-term (ST)- is associated with impaired haematopoiesis in the elderly3–6. Despite HSCs (Lin2c-Kit1Sca-12CD341Flk22 cells) (Extended Data Fig. 1h) a large amount of data describing the decline of HSC function on showed similar levels of b-catenin upon ageing (Fig. 1g and Extended ageing, the molecular mechanisms of this process remain largely Data Fig. 1i). Axin2 (an established direct downstream target of cano- unknown, which precludes rational approaches to attenuate stem- nical Wnt signalling9,15) transcript levels in aged LT-HSCs were mark- cell ageing. -

Novel Missense Mutations in the AXIN2 Gene Associated with Non
a r c h i v e s o f o r a l b i o l o g y 5 9 ( 2 0 1 4 ) 3 4 9 – 3 5 3 Available online at www.sciencedirect.com ScienceDirect journal homepage: http://www.elsevier.com/locate/aob Novel missense mutations in the AXIN2 gene associated with non-syndromic oligodontia a,1 a,1 b a Singwai Wong , Haochen Liu , Baojing Bai , Huaiguang Chang , c,d e a, a, Hongshan Zhao , Yixiang Wang , Dong Han **, Hailan Feng * a Department of Prosthodontics, Peking University School and Hospital of Stomatology, Beijing, China b Department of Prosthodontics, Beijing Stomatology Hospital, Beijing, China c Department of Medical Genetics, Peking University Health Science Center, Beijing, China d Peking University Center for Human Disease Genomics, Peking University Health Science Center, Beijing, China e Central Laboratory, Peking University School and Hospital of Stomatology, Beijing, China a r t i c l e i n f o a b s t r a c t Article history: Objective: Oligodontia, which is the congenital absence of six or more permanent teeth Accepted 23 December 2013 excluding third molars, may contribute to masticatory dysfunction, speech alteration, aesthetic problems and malocclusion. To date, mutations in EDA, AXIN2, MSX1, PAX9, Keywords: WNT10A, EDAR, EDARADD, NEMO and KRT 17 are known to associate with non-syndromic oligodontia. The aim of the study was to search for AXIN2 mutations in 96 patients with non- Non-syndromic oligodontia syndromic oligodontia. Mutation screening AXIN2 Design: We performed mutation analysis of 10 exons of the AXIN2 gene in 96 patients with isolated non-syndromic oligodontia. -

The WNT10A Gene in Ectodermal Dysplasias and Selective Tooth Agenesis Gabriele Mues,1* John Bonds,1 Lilin Xiang,1 Alexandre R
RESEARCH ARTICLE The WNT10A Gene in Ectodermal Dysplasias and Selective Tooth Agenesis Gabriele Mues,1* John Bonds,1 Lilin Xiang,1 Alexandre R. Vieira,2 Figen Seymen,3 Ophir Klein,4,5,6 and Rena N. D’Souza1 1Department of Biomedical Sciences, Texas A&M University-HSC Baylor College of Dentistry, Dallas, Texas 2Department of Oral Biology, University of Pittsburgh School of Dental Medicine, Pittsburgh, Pennsylvania 3University of Istanbul Faculty of Dentistry, Istanbul, Turkey 4Department of Orofacial Sciences, University of California San Francisco, San Francisco, California 5Department of Pediatrics, University of California San Francisco, San Francisco, California 6Institute for Human Genetics and Regeneration Medicine, University of California San Francisco, San Francisco, California Manuscript Received: 8 October 2013; Manuscript Accepted: 30 January 2014 Mutations in the WNT10A gene were first detected in the rare syndrome odonto-onycho-dermal dysplasia (OODD, How to Cite this Article: OMIM257980) but have now also been found to cause about Mues G, Bonds J, Xiang L, Vieira AR, 35–50% of selective tooth agenesis (STHAG4, OMIM150400), a Seymen F, Klein O, D’Souza RN. 2014. common disorder that mostly affects the permanent dentition. The WNT10A gene in ectodermal In our random sample of tooth agenesis patients, 40% had at least dysplasias and selective tooth agenesis. one mutation in the WNT10A gene. The WNT10A Phe228Ile variant alone reached an allele frequency of 0.21 in the tooth Am J Med Genet Part A 164A:2455–2460. agenesis cohort, about 10 times higher than the allele frequency reported in large SNP databases for Caucasian populations. Patients with bi-allelic WNT10A mutations have severe tooth syndrome (OMIM 224750) which additionally features eyelid cysts agenesis while heterozygous individuals are either unaffected or and predisposition to adnexal skin tumors. -
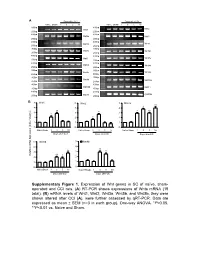
Supplementary Figure 1. Expression of Wnt Genes in SC of Naïve
A Days after CCI Days after CCI Naive Sham 1 3 5 10 Naive Sham 1 3 5 10 500bp 400bp Wnt1 Wnt2 250bp 250bp 400bp 400bp Wnt2b Wnt3 250bp 250bp 400bp 400bp Wnt3a Wnt4 250bp 250bp 300bp 400bp Wnt5a Wnt5b 200bp 250bp 400bp 400bp Wnt6 Wnt7a 250bp 250bp 400bp 400bp Wnt7b Wnt8a 250bp 250bp 400bp 350bp Wnt8b Wnt9a 250bp 200bp 400bp 400bp Wnt9b Wnt10a 250bp 250bp 400bp 400bp Wnt10b Wnt11 250bp 250bp 400bp 400bp Wnt16 GAPDH 250bp 250bp B 5 Wnt1 5 Wnt2 5 Wnt3a ** 4 4 4 * ** ** 3 3 * 3 * * 2 2 * 2 1 1 1 0 0 0 NaïveSham 1 3 5 10 Naïve Sham 1 3 5 10 Naïve Sham 1 3 5 10 Days after CCI Days after CCI Days after CCI 5 Wnt5b 5 Wnt8b 4 4 ** 3 ** 3 ** Relative mRNA Expression (fold of change) ** 2 * 2 1 1 0 0 NaïveSham 1 3 5 10 NaïveSham 1 3 5 10 Days after CCI Days after CCI Supplementary Figure 1 A B Negative Weak Moderate Strong Negative Weak Moderate Strong Large-sized cells 100% 100% 80% 80% 60% 60% 40% 20% 40% 0% 20% Sham CCI-1d CCI-14d Medium-sized cells Wnt3a immunoreactivity cells 0% 100% CGRP(+) IB4(+) 80% Small cells 60% 40% 20% 0% Sham CCI-1d CCI-14d Wnt3a immunoreactivity cells Small cells 100% 80% 60% 40% 20% 0% Sham CCI-1d CCI-14d Fig. S2 4 Fz1 4 Fz3 4 Fz4 4 Fz5 ** 3 3 ** 3 3 2 ** 2 2 ** 2 * 1 1 1 1 0 0 0 0 NaïveSham 1 5 10 NaïveSham 1 5 10 Naïve Sham 1 5 10 NaïveSham 1 5 10 Days after CCI Days after CCI Days after CCI Days after CCI 4 Fz6 4 Fz7 4 Fz8 4 Fz9 (Fold of Change) of (Fold 3 3 3 ** 3 Relative mRNA Expression Expression mRNA Relative ** 2 2 2 ** 2 * 1 1 1 1 0 0 0 0 NaïveSham 1 5 10 NaïveSham 1 5 10 Naïve Sham 1d 5d 10d NaïveSham 1